A virtual screen discovers novel, fragment-sized inhibitors of Mycobacterium tuberculosis InhA
- PMID: 25636146
- PMCID: PMC4386068
- DOI: 10.1021/ci500672v
A virtual screen discovers novel, fragment-sized inhibitors of Mycobacterium tuberculosis InhA
Abstract
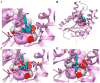
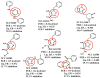
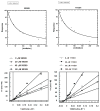
Isoniazid (INH) is usually administered to treat latent Mycobacterium tuberculosis (Mtb) infections and is used in combination therapy to treat active tuberculosis (TB). Unfortunately, resistance to this drug is hampering its clinical effectiveness. INH is a prodrug that must be activated by Mtb catalase-peroxidase (KatG) before it can inhibit InhA (Mtb enoyl-acyl-carrier-protein reductase). Isoniazid-resistant cases of TB found in clinical settings usually involve mutations in or deletion of katG, which abrogate INH activation. Compounds that inhibit InhA without requiring prior activation by KatG would not be affected by this resistance mechanism and hence would display continued potency against these drug-resistant isolates of Mtb. Virtual screening experiments versus InhA in the GO Fight Against Malaria (GO FAM) project were designed to discover new scaffolds that display base-stacking interactions with the NAD cofactor. GO FAM experiments included targets from other pathogens, including Mtb, when they had structural similarity to a malaria target. Eight of the 16 soluble compounds identified by docking against InhA plus visual inspection were modest inhibitors and did not require prior activation by KatG. The best two inhibitors discovered are both fragment-sized compounds and displayed Ki values of 54 and 59 μM, respectively. Importantly, the novel inhibitors discovered have low structural similarity to known InhA inhibitors and thus help expand the number of chemotypes on which future medicinal chemistry efforts can be focused. These new fragment hits could eventually help advance the fight against INH-resistant Mtb strains, which pose a significant global health threat.
Conflict of interest statement
The authors declare that they have no competing financial interest.
Figures



Similar articles
-
Identification of potential inhibitors of Mtb InhA: a pharmacoinformatics approach.J Biomol Struct Dyn. 2024 Sep;42(15):7957-7971. doi: 10.1080/07391102.2023.2242499. Epub 2023 Aug 1. J Biomol Struct Dyn. 2024. PMID: 37526169
-
Recent Advances and Structural Features of Enoyl-ACP Reductase Inhibitors of Mycobacterium tuberculosis.Arch Pharm (Weinheim). 2016 Nov;349(11):817-826. doi: 10.1002/ardp.201600186. Epub 2016 Oct 24. Arch Pharm (Weinheim). 2016. PMID: 27775177 Review.
-
The isoniazid-NAD adduct is a slow, tight-binding inhibitor of InhA, the Mycobacterium tuberculosis enoyl reductase: adduct affinity and drug resistance.Proc Natl Acad Sci U S A. 2003 Nov 25;100(24):13881-6. doi: 10.1073/pnas.2235848100. Epub 2003 Nov 17. Proc Natl Acad Sci U S A. 2003. PMID: 14623976 Free PMC article.
-
High affinity InhA inhibitors with activity against drug-resistant strains of Mycobacterium tuberculosis.ACS Chem Biol. 2006 Feb 17;1(1):43-53. doi: 10.1021/cb0500042. ACS Chem Biol. 2006. PMID: 17163639
-
Development of modern InhA inhibitors to combat drug resistant strains of Mycobacterium tuberculosis.Curr Top Med Chem. 2007;7(5):489-98. doi: 10.2174/156802607780059781. Curr Top Med Chem. 2007. PMID: 17346194 Review.
Cited by
-
Structure-based in silico approaches for drug discovery against Mycobacterium tuberculosis.Comput Struct Biotechnol J. 2021 Jun 24;19:3708-3719. doi: 10.1016/j.csbj.2021.06.034. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34285773 Free PMC article. Review.
-
Charting a Path to Success in Virtual Screening.Molecules. 2015 Oct 15;20(10):18732-58. doi: 10.3390/molecules201018732. Molecules. 2015. PMID: 26501243 Free PMC article. Review.
-
Predicting Mouse Liver Microsomal Stability with "Pruned" Machine Learning Models and Public Data.Pharm Res. 2016 Feb;33(2):433-49. doi: 10.1007/s11095-015-1800-5. Epub 2015 Sep 28. Pharm Res. 2016. PMID: 26415647 Free PMC article.
-
Intrabacterial Metabolism Obscures the Successful Prediction of an InhA Inhibitor of Mycobacterium tuberculosis.ACS Infect Dis. 2019 Dec 13;5(12):2148-2163. doi: 10.1021/acsinfecdis.9b00295. Epub 2019 Nov 5. ACS Infect Dis. 2019. PMID: 31625383 Free PMC article.
-
1001 Ways to run AutoDock Vina for virtual screening.J Comput Aided Mol Des. 2016 Mar;30(3):237-49. doi: 10.1007/s10822-016-9900-9. Epub 2016 Feb 20. J Comput Aided Mol Des. 2016. PMID: 26897747 Free PMC article.
References
-
- WHO. Global Tuberculosis Report. World Health Organization; Geneva: 2013.
-
- Abubakar I, Zignol M, Falzon D, Raviglione M, Ditiu L, Masham S, Adetifa I, Ford N, Cox H, Lawn SD, Marais BJ, McHugh TD, Mwaba P, Bates M, Lipman M, Zijenah L, Logan S, McNerney R, Zumla A, Sarda K, Nahid P, Hoelscher M, Pletschette M, Memish ZA, Kim P, Hafner R, Cole S, Migliori GB, Maeurer M, Schito M, Zumla A. Drug-Resistant Tuberculosis: Time for Visionary Political Leadership. The Lancet Infectious Diseases. 2013;13:529–539. - PubMed
-
- Dheda K, Migliori GB. The Global Rise of Extensively Drug-Resistant Tuberculosis: Is the Time to Bring Back Sanatoria Now Overdue? The Lancet. 2012;379:773–775. - PubMed
-
- Gothi D, Joshi JM. Resistant Tb: Newer Drugs and Community Approach. Recent Pat Antiinfect Drug Discovery. 2011;6:27–37. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

