A novel psychrophilic alkaline phosphatase from the metagenome of tidal flat sediments
- PMID: 25636680
- PMCID: PMC4335783
- DOI: 10.1186/s12896-015-0115-2
A novel psychrophilic alkaline phosphatase from the metagenome of tidal flat sediments
Abstract
Background: Alkaline phosphatase (AP) catalyzes the hydrolytic cleavage of phosphate monoesters under alkaline conditions and plays important roles in microbial ecology and molecular biology applications. Here, we report on the first isolation and biochemical characterization of a thermolabile AP from a metagenome.
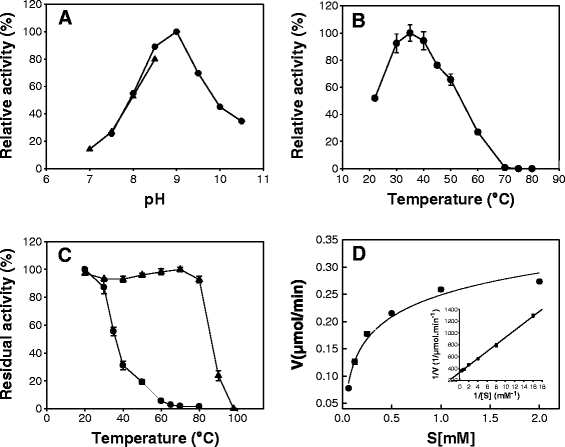
Results: The gene encoding a novel AP was isolated from a metagenomic library constructed with ocean-tidal flat sediments from the west coast of Korea. The metagenome-derived AP (mAP) gene composed of 1,824 nucleotides encodes a polypeptide with a calculated molecular mass of 64 kDa. The deduced amino acid sequence of mAP showed a high degree of similarity to other members of the AP family. Phylogenetic analysis revealed that the mAP is shown to be a member of a recently identified family of PhoX that is distinct from the well-studied classical PhoA family. When the open reading frame encoding mAP was cloned and expressed in recombinant Escherichia coli, the mature mAP was secreted to the periplasm and lacks an 81-amino-acid N-terminal Tat signal peptide. Mature mAP was purified to homogeneity as a monomeric enzyme with a molecular mass of 56 kDa. The purified mAP displayed typical features of a psychrophilic enzyme: high catalytic activity at low temperature and a remarkable thermal instability. The optimal temperature for the enzymatic activity of mAP was 37°C and complete thermal inactivation of the enzyme was observed at 65°C within 15 min. mAP was activated by Ca(2+) and exhibited maximal activity at pH 9.0. Except for phytic acid and glucose 1-phosphate, mAP showed phosphatase activity against various phosphorylated substrates indicating that it had low substrate specificity. In addition, the mAP was able to remove terminal phosphates from cohesive and blunt ends of linearized plasmid DNA, exhibiting comparable efficiency to commercially available APs that have been used in molecular biology.
Conclusions: The presented mAP enzyme is the first thermolabile AP found in cold-adapted marine metagenomes and may be useful for efficient dephosphorylation of linearized DNA.
Figures





References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

