Intestinal microbiota-derived metabolomic blood plasma markers for prior radiation injury
- PMID: 25636760
- PMCID: PMC4312583
- DOI: 10.1016/j.ijrobp.2014.10.023
Intestinal microbiota-derived metabolomic blood plasma markers for prior radiation injury
Abstract
Purpose: Assessing whole-body radiation injury and absorbed dose is essential for remediation efforts following accidental or deliberate exposure in medical, industrial, military, or terrorist incidents. We hypothesize that variations in specific metabolite concentrations extracted from blood plasma would correlate with whole-body radiation injury and dose.
Methods and materials: Groups of C57BL/6 mice (n=12 per group) were exposed to 0, 2, 4, 8, and 10.4 Gy of whole-body gamma radiation. At 24 hours after treatment, all animals were euthanized, and both plasma and liver biopsy samples were obtained, the latter being used to identify a distinct hepatic radiation injury response within plasma. A semiquantitative, untargeted metabolite/lipid profile was developed using gas chromatography-mass spectrometry and liquid chromatography-tandem mass spectrometry, which identified 354 biochemical compounds. A second set of C57BL/6 mice (n=6 per group) were used to assess a subset of identified plasma markers beyond 24 hours.
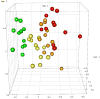
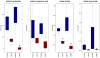
Results: We identified a cohort of 37 biochemical compounds in plasma that yielded the optimal separation of the irradiated sample groups, with the most correlated metabolites associated with pyrimidine (positively correlated) and tryptophan (negatively correlated) metabolism. The latter were predominantly associated with indole compounds, and there was evidence that these were also correlated between liver and plasma. No evidence of saturation as a function of dose was observed, as has been noted for studies involving metabolite analysis of urine.
Conclusions: Plasma profiling of specific metabolites related to pyrimidine and tryptophan pathways can be used to differentiate whole-body radiation injury and dose response. As the tryptophan-associated indole compounds have their origin in the intestinal microbiome and subsequently the liver, these metabolites particularly represent an attractive marker for radiation injury within blood plasma.
Copyright © 2015 Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures

References
-
- Stone HB, Coleman CN, Anscher MS, McBride WH. Effects of radiation on normal tissue: Consequences and mechanisms. The lancet oncology. 2003;4:529–536. - PubMed
-
- Friesecke I, Beyrer K, Fliedner TM system MtMtpfravaabfacg. How to cope with radiation accidents: The medical management. Br J Radiol. 2001;74:121–122. - PubMed
-
- Fiehn O. Metabolomics--the link between genotypes and phenotypes. Plant molecular biology. 2002;48:155–171. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

