The genetics of splicing in neuroblastoma
- PMID: 25637275
- PMCID: PMC4390477
- DOI: 10.1158/2159-8290.CD-14-0892
The genetics of splicing in neuroblastoma
Abstract
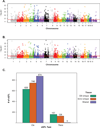
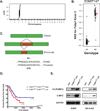
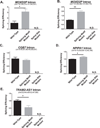
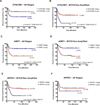
Regulation of mRNA splicing, a critical and tightly regulated cellular function, underlies the majority of proteomic diversity and is frequently disrupted in disease. Using an integrative genomics approach, we combined both genomic data and exon-level transcriptome data in two somatic tissues (cerebella and peripheral ganglia) from a transgenic mouse model of neuroblastoma, a tumor that arises from the peripheral neural crest. Here, we describe splicing quantitative trait loci associated with differential splicing across the genome that we use to identify genes with previously unknown functions within the splicing pathway and to define de novo intronic splicing motifs that influence splicing from hundreds of bases away. Our results show that these splicing motifs represent sites for functional recurrent mutations and highlight novel candidate genes in human cancers, including childhood neuroblastoma.
Significance: Somatic mutations with predictable downstream effects are largely relegated to coding regions, which comprise less than 2% of the human genome. Using an unbiased in vivo analysis of a mouse model of neuroblastoma, we have identified intronic splicing motifs that translate into sites for recurrent somatic mutations in human cancers.
©2015 American Association for Cancer Research.
Conflict of interest statement
The authors do not have any conflicts of interest.
Figures


References
-
- Garcia-Blanco M, Baraniak A, Lasda E, Baraniak E. Nat Biotechnol. Vol. 22. Nature Publishing Group; 2004. Alternative splicing in disease and therapy; pp. 535–546. - PubMed
-
- Chen J, Weiss WA. Oncogene. Macmillan Publishers Limited; 2014. Alternative splicing in cancer: implications for biology and therapy. - PubMed
-
- Brooks AN, Choi PS, de Waal L, Sharifnia T, Imielinski M, Saksena G, et al. A Pan-Cancer Analysis of Transcriptome Changes Associated with Somatic Mutations in U2AF1 Reveals Commonly Altered Splicing Events. In: Ast G, editor. PLoS One. Vol. 9. Public Library of Science; 2014. p. e87361. - PMC - PubMed
-
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K08 NS079485/NS/NINDS NIH HHS/United States
- R01 CA102321/CA/NCI NIH HHS/United States
- U01 CA176287/CA/NCI NIH HHS/United States
- CA159859/CA/NCI NIH HHS/United States
- CA82103/CA/NCI NIH HHS/United States
- 5P30CA082103-15/CA/NCI NIH HHS/United States
- CA176287/CA/NCI NIH HHS/United States
- P01 CA081403/CA/NCI NIH HHS/United States
- R01 CA163336/CA/NCI NIH HHS/United States
- R01 CA159859/CA/NCI NIH HHS/United States
- P30 CA082103/CA/NCI NIH HHS/United States
- CA148699/CA/NCI NIH HHS/United States
- CA102321/CA/NCI NIH HHS/United States
- CA081403/CA/NCI NIH HHS/United States
- R01 CA148699/CA/NCI NIH HHS/United States
- T32 GM008568/GM/NIGMS NIH HHS/United States
- R01CA163336/CA/NCI NIH HHS/United States
- T32 CA128583/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

