A whole-genome shotgun approach for assembling and anchoring the hexaploid bread wheat genome
- PMID: 25637298
- PMCID: PMC4373400
- DOI: 10.1186/s13059-015-0582-8
A whole-genome shotgun approach for assembling and anchoring the hexaploid bread wheat genome
Abstract
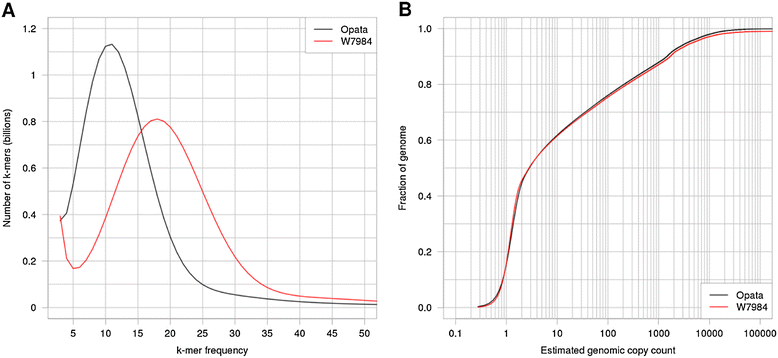
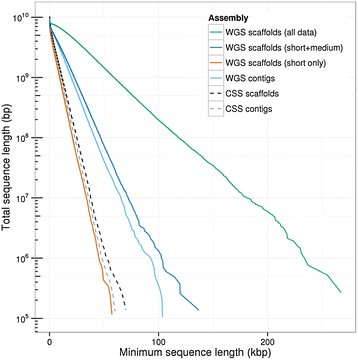
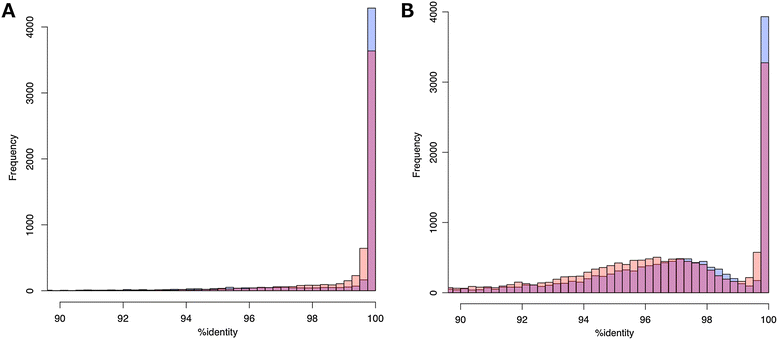
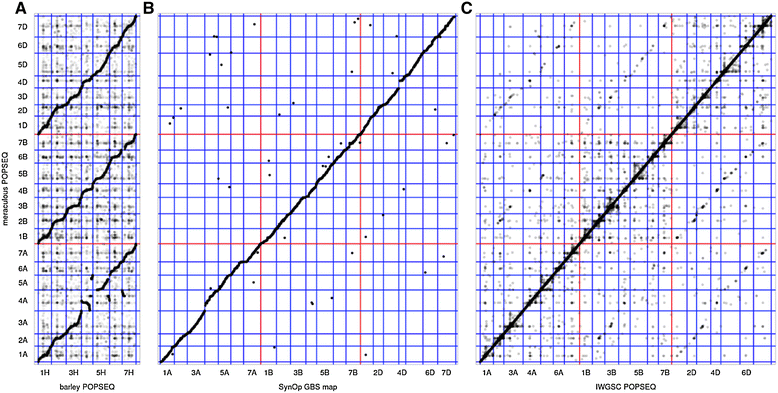
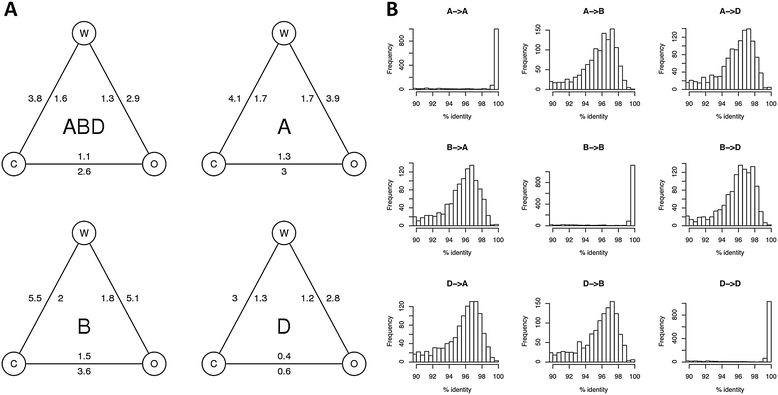
Polyploid species have long been thought to be recalcitrant to whole-genome assembly. By combining high-throughput sequencing, recent developments in parallel computing, and genetic mapping, we derive, de novo, a sequence assembly representing 9.1 Gbp of the highly repetitive 16 Gbp genome of hexaploid wheat, Triticum aestivum, and assign 7.1 Gb of this assembly to chromosomal locations. The genome representation and accuracy of our assembly is comparable or even exceeds that of a chromosome-by-chromosome shotgun assembly. Our assembly and mapping strategy uses only short read sequencing technology and is applicable to any species where it is possible to construct a mapping population.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

