Superiority of transcriptional profiling over procalcitonin for distinguishing bacterial from viral lower respiratory tract infections in hospitalized adults
- PMID: 25637350
- PMCID: PMC4565998
- DOI: 10.1093/infdis/jiv047
Superiority of transcriptional profiling over procalcitonin for distinguishing bacterial from viral lower respiratory tract infections in hospitalized adults
Erratum in
-
Suarez et al (J Infect Dis 2015; 212:213-22).J Infect Dis. 2015 Dec 15;212(12):2023. doi: 10.1093/infdis/jiv514. J Infect Dis. 2015. PMID: 26598315 Free PMC article. No abstract available.
Abstract
Background: Distinguishing between bacterial and viral lower respiratory tract infection (LRTI) remains challenging. Transcriptional profiling is a promising tool for improving diagnosis in LRTI.
Methods: We performed whole blood transcriptional analysis in 118 patients (median age [interquartile range], 61 [50-76] years) hospitalized with LRTI and 40 age-matched healthy controls (median age, 60 [46-70] years). We applied class comparisons, modular analysis, and class prediction algorithms to identify and validate diagnostic biosignatures for bacterial and viral LRTI.
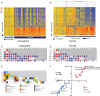
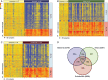
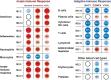
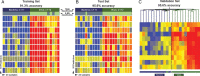
Results: Patients were classified as having bacterial (n = 22), viral (n = 71), or bacterial-viral LRTI (n = 25) based on comprehensive microbiologic testing. Compared with healthy controls, statistical group comparisons (P < .01; multiple-test corrections) identified 3376 differentially expressed genes in patients with bacterial LRTI, 2391 in viral LRTI, and 2628 in bacterial-viral LRTI. Patients with bacterial LRTI showed significant overexpression of inflammation and neutrophil genes (bacterial > bacterial-viral > viral), and those with viral LRTI displayed significantly greater overexpression of interferon genes (viral > bacterial-viral > bacterial). The K-nearest neighbors algorithm identified 10 classifier genes that discriminated between bacterial and viral LRTI with a 95% sensitivity (95% confidence interval, 77%-100%) and 92% specificity (77%-98%), compared with a sensitivity of 38% (18%-62%) and a specificity of 91% (76%-98%) for procalcitonin.
Conclusions: Transcriptional profiling is a helpful tool for diagnosis of LRTI.
Keywords: bacterial infections; lower respiratory tract infection; microarrays; procalcitonin; viral infections.
© The Author 2015. Published by Oxford University Press on behalf of the Infectious Diseases Society of America. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures
Comment in
-
Moving toward prime time: host signatures for diagnosis of respiratory infections.J Infect Dis. 2015 Jul 15;212(2):173-5. doi: 10.1093/infdis/jiv032. Epub 2015 Jan 29. J Infect Dis. 2015. PMID: 25637349 No abstract available.
Similar articles
-
The utility of biomarkers in differentiating bacterial from non-bacterial lower respiratory tract infection in hospitalized children: difference of the diagnostic performance between acute pneumonia and bronchitis.J Infect Chemother. 2014 Oct;20(10):616-20. doi: 10.1016/j.jiac.2014.06.003. Epub 2014 Jul 11. J Infect Chemother. 2014. PMID: 25027057
-
Serum Procalcitonin Measurement and Viral Testing to Guide Antibiotic Use for Respiratory Infections in Hospitalized Adults: A Randomized Controlled Trial.J Infect Dis. 2015 Dec 1;212(11):1692-700. doi: 10.1093/infdis/jiv252. Epub 2015 Apr 24. J Infect Dis. 2015. PMID: 25910632 Free PMC article. Clinical Trial.
-
Value of serum procalcitonin, neopterin, and C-reactive protein in differentiating bacterial from viral etiologies in patients presenting with lower respiratory tract infections.Diagn Microbiol Infect Dis. 2007 Oct;59(2):131-6. doi: 10.1016/j.diagmicrobio.2007.04.019. Epub 2007 Jul 26. Diagn Microbiol Infect Dis. 2007. PMID: 17662560
-
What cardiologists do need to know about procalcitonin.Clin Lab. 2005;51(1-2):1-4. Clin Lab. 2005. PMID: 15719698 Review.
-
Procalcitonin for triage of patients with respiratory tract symptoms: a case study in the trial design process for approval of a new diagnostic test for lower respiratory tract infections.Clin Infect Dis. 2011 May;52 Suppl 4:S351-6. doi: 10.1093/cid/cir058. Clin Infect Dis. 2011. PMID: 21460295 Review.
Cited by
-
Benchmarking transcriptional host response signatures for infection diagnosis.Cell Syst. 2022 Dec 21;13(12):974-988.e7. doi: 10.1016/j.cels.2022.11.007. Cell Syst. 2022. PMID: 36549274 Free PMC article.
-
Rhinovirus Detection in Symptomatic and Asymptomatic Children: Value of Host Transcriptome Analysis.Am J Respir Crit Care Med. 2016 Apr 1;193(7):772-82. doi: 10.1164/rccm.201504-0749OC. Am J Respir Crit Care Med. 2016. PMID: 26571305 Free PMC article.
-
Respiratory viral infections are underdiagnosed in patients with suspected sepsis.Eur J Clin Microbiol Infect Dis. 2017 Oct;36(10):1767-1776. doi: 10.1007/s10096-017-2990-z. Epub 2017 May 17. Eur J Clin Microbiol Infect Dis. 2017. PMID: 28516200 Free PMC article.
-
Antiviral Response in the Nasopharynx Identifies Patients With Respiratory Virus Infection.J Infect Dis. 2018 Mar 5;217(6):897-905. doi: 10.1093/infdis/jix648. J Infect Dis. 2018. PMID: 29281100 Free PMC article.
-
A blood-based host gene expression assay for early detection of respiratory viral infection: an index-cluster prospective cohort study.Lancet Infect Dis. 2021 Mar;21(3):396-404. doi: 10.1016/S1473-3099(20)30486-2. Epub 2020 Sep 24. Lancet Infect Dis. 2021. PMID: 32979932 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

