RAMPART: a workflow management system for de novo genome assembly
- PMID: 25637556
- PMCID: PMC4443680
- DOI: 10.1093/bioinformatics/btv056
RAMPART: a workflow management system for de novo genome assembly
Abstract
Motivation: The de novo assembly of genomes from whole- genome shotgun sequence data is a computationally intensive, multi-stage task and it is not known a priori which methods and parameter settings will produce optimal results. In current de novo assembly projects, a popular strategy involves trying many approaches, using different tools and settings, and then comparing and contrasting the results in order to select a final assembly for publication.
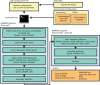
Results: Herein, we present RAMPART, a configurable workflow management system for de novo genome assembly, which helps the user identify combinations of third-party tools and settings that provide good results for their particular genome and sequenced reads. RAMPART is designed to exploit High performance computing environments, such as clusters and shared memory systems, where available.
Availability and implementation: RAMPART is available under the GPLv3 license at: https://github.com/TGAC/RAMPART.
© The Author 2015. Published by Oxford University Press.
Figures
References
-
- Seemann T. (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics , 30, 2068–2069. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

