Molecular underpinnings of Aprataxin RNA/DNA deadenylase function and dysfunction in neurological disease
- PMID: 25637650
- PMCID: PMC4417397
- DOI: 10.1016/j.pbiomolbio.2015.01.007
Molecular underpinnings of Aprataxin RNA/DNA deadenylase function and dysfunction in neurological disease
Abstract
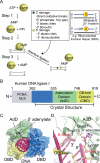
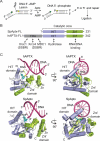
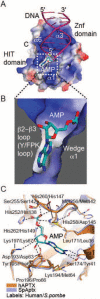
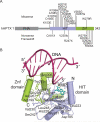
Eukaryotic DNA ligases seal DNA breaks in the final step of DNA replication and repair transactions via a three-step reaction mechanism that can abort if DNA ligases encounter modified DNA termini, such as the products and repair intermediates of DNA oxidation, alkylation, or the aberrant incorporation of ribonucleotides into genomic DNA. Such abortive DNA ligation reactions act as molecular checkpoint for DNA damage and create 5'-adenylated nucleic acid termini in the context of DNA and RNA-DNA substrates in DNA single strand break repair (SSBR) and ribonucleotide excision repair (RER). Aprataxin (APTX), a protein altered in the heritable neurological disorder Ataxia with Oculomotor Apraxia 1 (AOA1), acts as a DNA ligase "proofreader" to directly reverse AMP-modified nucleic acid termini in DNA- and RNA-DNA damage responses. Herein, we survey APTX function and the emerging cell biological, structural and biochemical data that has established a molecular foundation for understanding the APTX mediated deadenylation reaction, and is providing insights into the molecular bases of APTX deficiency in AOA1.
Keywords: AOA1; Aprataxin; Aptx; DNA damage response; DNA ligase; Neurodegenerative disease.
Published by Elsevier Ltd.
Figures


References
-
- Ahel I, Rass U, El-Khamisy SF, Katyal S, Clements PM, McKinnon PJ, Caldecott KW, West SC. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 2006;443:713–716. - PubMed
-
- Aicardi J, Barbosa C, Andermann E, Andermann F, Morcos R, Ghanem Q, Fukuyama Y, Awaya Y, Moe P. Ataxia-ocular motor apraxia: a syndrome mimicking ataxia-telangiectasia. Ann. Neurol. 1988;24:497–502. - PubMed
-
- Baba Y, Uitti RJ, Boylan KB, Uehara Y, Yamada T, Farrer MJ, Couchon E, Batish SD, Wszolek ZK. Aprataxin (APTX) gene mutations resembling multiple system atrophy. Parkinsonism Relat. Disord. 2007;13:139–142. - PubMed
-
- Becherel OJ, Gueven N, Birrell GW, Schreiber V, Suraweera A, Jakob B, Taucher-Scholz G, Lavin MF. Nucleolar localization of aprataxin is dependent on interaction with nucleolin and on active ribosomal DNA transcription. Hum Mol Genet. 2006;15:2239–2249. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
