A micropeptide encoded by a putative long noncoding RNA regulates muscle performance
- PMID: 25640239
- PMCID: PMC4356254
- DOI: 10.1016/j.cell.2015.01.009
A micropeptide encoded by a putative long noncoding RNA regulates muscle performance
Abstract
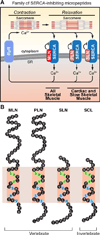
Functional micropeptides can be concealed within RNAs that appear to be noncoding. We discovered a conserved micropeptide, which we named myoregulin (MLN), encoded by a skeletal muscle-specific RNA annotated as a putative long noncoding RNA. MLN shares structural and functional similarity with phospholamban (PLN) and sarcolipin (SLN), which inhibit SERCA, the membrane pump that controls muscle relaxation by regulating Ca(2+) uptake into the sarcoplasmic reticulum (SR). MLN interacts directly with SERCA and impedes Ca(2+) uptake into the SR. In contrast to PLN and SLN, which are expressed in cardiac and slow skeletal muscle in mice, MLN is robustly expressed in all skeletal muscle. Genetic deletion of MLN in mice enhances Ca(2+) handling in skeletal muscle and improves exercise performance. These findings identify MLN as an important regulator of skeletal muscle physiology and highlight the possibility that additional micropeptides are encoded in the many RNAs currently annotated as noncoding.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures


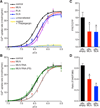
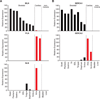
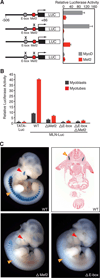
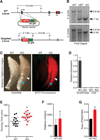
Comment in
-
One small step for muscle: a new micropeptide regulates performance.Cell Metab. 2015 Apr 7;21(4):515-6. doi: 10.1016/j.cmet.2015.03.013. Cell Metab. 2015. PMID: 25863244
References
-
- Allen DG, Gervasio OL, Yeung EW, Whitehead NP. Calcium and the damage pathways in muscular dystrophy. Can J Physiol Pharmacol. 2010;88:83–91. - PubMed
-
- Anderson DM, Beres BJ, Wilson-Rawls J, Rawls A. The homeobox gene Mohawk represses transcription by recruiting the sin3A/HDAC co-repressor complex. Dev Dyn. 2009;238:572–580. - PubMed
-
- Andrews SJ, Rothnagel JA. Emerging evidence for functional peptides encoded by short open reading frames. Nat Rev Genet. 2014;15:193–204. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL077439/HL/NHLBI NIH HHS/United States
- R01 DK099653/DK/NIDDK NIH HHS/United States
- DK-099653/DK/NIDDK NIH HHS/United States
- U01 HL100401/HL/NHLBI NIH HHS/United States
- R01 HL053351/HL/NHLBI NIH HHS/United States
- R01 HL111665/HL/NHLBI NIH HHS/United States
- HL-077439/HL/NHLBI NIH HHS/United States
- U01-HL-100401/HL/NHLBI NIH HHS/United States
- HL-111665/HL/NHLBI NIH HHS/United States
- HL-093039/HL/NHLBI NIH HHS/United States
- 1F30AR067094-01/AR/NIAMS NIH HHS/United States
- F30 AR067094/AR/NIAMS NIH HHS/United States
- R01 HL093039/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

