Next-generation sequencing of duplication CNVs reveals that most are tandem and some create fusion genes at breakpoints
- PMID: 25640679
- PMCID: PMC4320257
- DOI: 10.1016/j.ajhg.2014.12.017
Next-generation sequencing of duplication CNVs reveals that most are tandem and some create fusion genes at breakpoints
Abstract
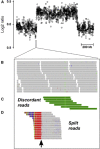
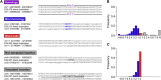
Interpreting the genomic and phenotypic consequences of copy-number variation (CNV) is essential to understanding the etiology of genetic disorders. Whereas deletion CNVs lead obviously to haploinsufficiency, duplications might cause disease through triplosensitivity, gene disruption, or gene fusion at breakpoints. The mutational spectrum of duplications has been studied at certain loci, and in some cases these copy-number gains are complex chromosome rearrangements involving triplications and/or inversions. However, the organization of clinically relevant duplications throughout the genome has yet to be investigated on a large scale. Here we fine-mapped 184 germline duplications (14.7 kb-25.3 Mb; median 532 kb) ascertained from individuals referred for diagnostic cytogenetics testing. We performed next-generation sequencing (NGS) and whole-genome sequencing (WGS) to sequence 130 breakpoints from 112 subjects with 119 CNVs and found that most (83%) were tandem duplications in direct orientation. The remainder were triplications embedded within duplications (8.4%), adjacent duplications (4.2%), insertional translocations (2.5%), or other complex rearrangements (1.7%). Moreover, we predicted six in-frame fusion genes at sequenced duplication breakpoints; four gene fusions were formed by tandem duplications, one by two interconnected duplications, and one by duplication inserted at another locus. These unique fusion genes could be related to clinical phenotypes and warrant further study. Although most duplications are positioned head-to-tail adjacent to the original locus, those that are inverted, triplicated, or inserted can disrupt or fuse genes in a manner that might not be predicted by conventional copy-number assays. Therefore, interpreting the genetic consequences of duplication CNVs requires breakpoint-level analysis.
Copyright © 2015 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Kearney H.M., Thorland E.C., Brown K.K., Quintero-Rivera F., South S.T., Working Group of the American College of Medical Genetics Laboratory Quality Assurance Committee American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet. Med. 2011;13:680–685. - PubMed
-
- Cook E.H., Jr., Scherer S.W. Copy-number variations associated with neuropsychiatric conditions. Nature. 2008;455:919–923. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

