Establishing Chlamydomonas reinhardtii as an industrial biotechnology host
- PMID: 25641561
- PMCID: PMC4515103
- DOI: 10.1111/tpj.12781
Establishing Chlamydomonas reinhardtii as an industrial biotechnology host
Abstract
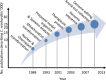
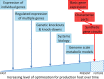
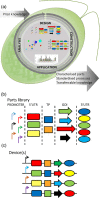
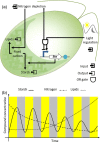
Microalgae constitute a diverse group of eukaryotic unicellular organisms that are of interest for pure and applied research. Owing to their natural synthesis of value-added natural products microalgae are emerging as a source of sustainable chemical compounds, proteins and metabolites, including but not limited to those that could replace compounds currently made from fossil fuels. For the model microalga, Chlamydomonas reinhardtii, this has prompted a period of rapid development so that this organism is poised for exploitation as an industrial biotechnology platform. The question now is how best to achieve this? Highly advanced industrial biotechnology systems using bacteria and yeasts were established in a classical metabolic engineering manner over several decades. However, the advent of advanced molecular tools and the rise of synthetic biology provide an opportunity to expedite the development of C. reinhardtii as an industrial biotechnology platform, avoiding the process of incremental improvement. In this review we describe the current status of genetic manipulation of C. reinhardtii for metabolic engineering. We then introduce several concepts that underpin synthetic biology, and show how generic parts are identified and used in a standard manner to achieve predictable outputs. Based on this we suggest that the development of C. reinhardtii as an industrial biotechnology platform can be achieved more efficiently through adoption of a synthetic biology approach.
Keywords: Chlamydomonas reinhardtii; industrial biotechnology; metabolic engineering; rational design; synthetic biology; transgene expression.
© 2015 The Authors The Plant Journal published by Society for Experimental Biology and John Wiley & Sons Ltd.
Figures
References
-
- Allmer J, Naumann B, Markert C, Zhang M, Hippler M. Mass spectrometric genomic data mining: novel insights into bioenergetic pathways in Chlamydomonas reinhardtii. Proteomics. 2006;6:6207–6220. - PubMed
-
- Anthony JR, Anthony LC, Nowroozi F, Kwon G, Newman JD, Keasling JD. Optimization of the mevalonate-based isoprenoid biosynthetic pathway in Escherichia coli for production of the anti-malarial drug precursor amorpha-4,11-diene. Metab. Eng. 2009;11:13–19. - PubMed
-
- Baltz A, Dang K-V, Beyly A, Auroy P, Richaud P, Cournac L, Peltier G. Plastidial expression of type II NAD(P)H dehydrogenase increases the reducing state of plastoquinones and hydrogen photoproduction rate by the indirect pathway in Chlamydomonas reinhardtii. Plant Physiol. 2014;165:1344–1352. - PMC - PubMed
-
- Barnes D, Franklin S, Schultz J, Henry R, Brown E, Coragliotti A, Mayfield SP. Contribution of 5'- and 3'-untranslated regions of plastid mRNAs to the expression of Chlamydomonas reinhardtii chloroplast genes. Mol. Genet. Genomics. 2005;274:625–636. - PubMed
Publication types
MeSH terms
Grants and funding
- BB/D011043/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/I013164/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/L014130/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/I00680X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

