Next-generation sequencing approach for connecting secondary metabolites to biosynthetic gene clusters in fungi
- PMID: 25642215
- PMCID: PMC4294208
- DOI: 10.3389/fmicb.2014.00774
Next-generation sequencing approach for connecting secondary metabolites to biosynthetic gene clusters in fungi
Abstract
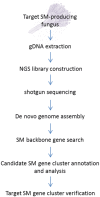
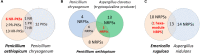
Genomics has revolutionized the research on fungal secondary metabolite (SM) biosynthesis. To elucidate the molecular and enzymatic mechanisms underlying the biosynthesis of a specific SM compound, the important first step is often to find the genes that responsible for its synthesis. The accessibility to fungal genome sequences allows the bypass of the cumbersome traditional library construction and screening approach. The advance in next-generation sequencing (NGS) technologies have further improved the speed and reduced the cost of microbial genome sequencing in the past few years, which has accelerated the research in this field. Here, we will present an example work flow for identifying the gene cluster encoding the biosynthesis of SMs of interest using an NGS approach. We will also review the different strategies that can be employed to pinpoint the targeted gene clusters rapidly by giving several examples stemming from our work.
Keywords: filamentous fungi; gene clusters; genome mining; next generation sequencing; secondary metabolites.
Figures



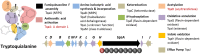

Similar articles
-
Strategies to establish the link between biosynthetic gene clusters and secondary metabolites.Fungal Genet Biol. 2019 Sep;130:107-121. doi: 10.1016/j.fgb.2019.06.001. Epub 2019 Jun 10. Fungal Genet Biol. 2019. PMID: 31195124 Review.
-
A chemical ecogenomics approach to understand the roles of secondary metabolites in fungal cereal pathogens.Front Microbiol. 2014 Nov 19;5:640. doi: 10.3389/fmicb.2014.00640. eCollection 2014. Front Microbiol. 2014. PMID: 25477876 Free PMC article.
-
Nonribosomal peptides in fungal cell factories: from genome mining to optimized heterologous production.Biotechnol Adv. 2019 Dec;37(8):107449. doi: 10.1016/j.biotechadv.2019.107449. Epub 2019 Sep 10. Biotechnol Adv. 2019. PMID: 31518630 Review.
-
Filamentous fungi from extreme environments as a promising source of novel bioactive secondary metabolites.Front Microbiol. 2015 Sep 9;6:903. doi: 10.3389/fmicb.2015.00903. eCollection 2015. Front Microbiol. 2015. PMID: 26441853 Free PMC article.
-
Recent advances in the genome mining of Aspergillus secondary metabolites (covering 2012-2018).Medchemcomm. 2019 Apr 26;10(6):840-866. doi: 10.1039/c9md00054b. eCollection 2019 Jun 1. Medchemcomm. 2019. PMID: 31303983 Free PMC article. Review.
Cited by
-
Boosting Secondary Metabolite Production and Discovery through the Engineering of Novel Microbial Biosensors.Biomed Res Int. 2018 Jul 9;2018:7021826. doi: 10.1155/2018/7021826. eCollection 2018. Biomed Res Int. 2018. PMID: 30079350 Free PMC article. Review.
-
Iso-Seq analysis of the Taxus cuspidata transcriptome reveals the complexity of Taxol biosynthesis.BMC Plant Biol. 2019 May 21;19(1):210. doi: 10.1186/s12870-019-1809-8. BMC Plant Biol. 2019. PMID: 31113367 Free PMC article.
-
Technology development for natural product biosynthesis in Saccharomyces cerevisiae.Curr Opin Biotechnol. 2016 Dec;42:74-83. doi: 10.1016/j.copbio.2016.02.033. Epub 2016 Mar 16. Curr Opin Biotechnol. 2016. PMID: 26994377 Free PMC article. Review.
-
High throughput, small scale methods to characterise the growth of marine fungi.PLoS One. 2020 Aug 7;15(8):e0236822. doi: 10.1371/journal.pone.0236822. eCollection 2020. PLoS One. 2020. PMID: 32764772 Free PMC article.
-
Synergistic Biocontrol and Growth Promotion in Strawberries by Co-Cultured Trichoderma harzianum TW21990 and Burkholderia vietnamiensis B418.J Fungi (Basel). 2024 Aug 5;10(8):551. doi: 10.3390/jof10080551. J Fungi (Basel). 2024. PMID: 39194877 Free PMC article.
References
-
- Adefarati A. A., Giacobbe R. A., Hensens O. D., Tkacz J. S. (1991). Biosynthesis of L-671,329, an echinocandin-type antibiotic produced by Zalerion arboricola: origins of some of the unusual amino acids and the dimethylmyristic acid side chain. J. Am. Chem. Soc. 113 3542–3545 10.1021/ja00009a048 - DOI
-
- Ali H., Ries M. I., Lankhorst P. P., Van Der Hoeven R. A., Schouten O. L., Noga M., et al. (2014). A non-canonical NRPS is involved in the synthesis of fungisporin and related hydrophobic cyclic tetrapeptides in Penicillium chrysogenum. PLoS ONE 9:e98212 10.1371/journal.pone.0098212 - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

