An MRPS12 mutation modifies aminoglycoside sensitivity caused by 12S rRNA mutations
- PMID: 25642242
- PMCID: PMC4294204
- DOI: 10.3389/fgene.2014.00469
An MRPS12 mutation modifies aminoglycoside sensitivity caused by 12S rRNA mutations
Abstract
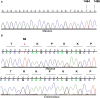
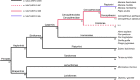
Several homoplasmic pathologic mutations in mitochondrial DNA, such as those causing Leber hereditary optic neuropathy or non-syndromic hearing loss, show incomplete penetrance. Therefore, other elements must modify their pathogenicity. Discovery of these modifying factors is not an easy task because in multifactorial diseases conventional genetic approaches may not always be informative. Here, we have taken an evolutionary approach to unmask putative modifying factors for a particular homoplasmic pathologic mutation causing aminoglycoside-induced and non-syndromic hearing loss, the m.1494C>T transition in the mitochondrial DNA. The mutation is located in the decoding site of the mitochondrial ribosomal RNA. We first looked at mammalian species that had fixed the human pathologic mutation. These mutations are called compensated pathogenic deviations because an organism carrying one must also have another that suppresses the deleterious effect of the first. We found that species from the primate family Cercopithecidae (old world monkeys) harbor the m.1494T allele even if their auditory function is normal. In humans the m.1494T allele increases the susceptibility to aminoglycosides. However, in primary fibroblasts from a Cercopithecidae species, aminoglycosides do not impair cell growth, respiratory complex IV activity and quantity or the mitochondrial protein synthesis. Interestingly, this species also carries a fixed mutation in the mitochondrial ribosomal protein S12. We show that the expression of this variant in a human m.1494T cell line reduces its susceptibility to aminoglycosides. Because several mutations in this human protein have been described, they may possibly explain the absence of pathologic phenotype in some pedigree members with the most frequent pathologic mutations in mitochondrial ribosomal RNA.
Keywords: aminoglycoside; compensatory mutation; evolutionary approaches; incomplete penetrance; mitochondrial ribosomal RNA; mitochondrial ribosomal protein S12; pathologic mutation.
Figures







Similar articles
-
Mitochondrial 12S rRNA mutations associated with aminoglycoside ototoxicity.Mitochondrion. 2011 Mar;11(2):237-45. doi: 10.1016/j.mito.2010.10.006. Epub 2010 Nov 1. Mitochondrion. 2011. PMID: 21047563 Review.
-
Mitochondrial 12S rRNA A827G mutation is involved in the genetic susceptibility to aminoglycoside ototoxicity.Biochem Biophys Res Commun. 2006 Aug 11;346(4):1131-5. doi: 10.1016/j.bbrc.2006.05.208. Epub 2006 Jun 12. Biochem Biophys Res Commun. 2006. PMID: 16782057
-
The contribution of the mitochondrial COI/tRNA(Ser(UCN)) gene mutations to non-syndromic and aminoglycoside-induced hearing loss in Polish patients.Mol Genet Metab. 2011 Sep-Oct;104(1-2):153-9. doi: 10.1016/j.ymgme.2011.05.004. Epub 2011 May 13. Mol Genet Metab. 2011. PMID: 21621438
-
A mutation in mitochondrial 12S rRNA, A827G, in Argentinean family with hearing loss after aminoglycoside treatment.Biochem Biophys Res Commun. 2008 Apr 11;368(3):631-6. doi: 10.1016/j.bbrc.2008.01.143. Epub 2008 Feb 7. Biochem Biophys Res Commun. 2008. PMID: 18261986
-
Genetics of aminoglycoside-induced and prelingual non-syndromic mitochondrial hearing impairment: a review.Int J Audiol. 2008 Nov;47(11):702-7. doi: 10.1080/14992020802215862. Int J Audiol. 2008. PMID: 19031229 Review.
Cited by
-
Mutations of the mitochondrial carrier translocase channel subunit TIM22 cause early-onset mitochondrial myopathy.Hum Mol Genet. 2018 Dec 1;27(23):4135-4144. doi: 10.1093/hmg/ddy305. Hum Mol Genet. 2018. PMID: 30452684 Free PMC article.
-
Development and characterization of cell models harbouring mtDNA deletions for in vitro study of Pearson syndrome.Dis Model Mech. 2022 Mar 1;15(3):dmm049083. doi: 10.1242/dmm.049083. Epub 2022 Mar 1. Dis Model Mech. 2022. PMID: 35191981 Free PMC article.
-
Infectious stress triggers a POLG-related mitochondrial disease.Neurogenetics. 2020 Jan;21(1):19-27. doi: 10.1007/s10048-019-00593-2. Epub 2019 Oct 26. Neurogenetics. 2020. PMID: 31655921
-
Is population frequency a useful criterion to assign pathogenicity to newly described mitochondrial DNA variants?Orphanet J Rare Dis. 2022 Aug 19;17(1):316. doi: 10.1186/s13023-022-02428-0. Orphanet J Rare Dis. 2022. PMID: 35986281 Free PMC article.
-
Mitochondrial dysfunction and its role in tissue-specific cellular stress.Cell Stress. 2018 Jul 13;2(8):184-199. doi: 10.15698/cst2018.07.147. Cell Stress. 2018. PMID: 31225486 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

