Inhibitor of DNA Binding 4 (ID4) is highly expressed in human melanoma tissues and may function to restrict normal differentiation of melanoma cells
- PMID: 25642713
- PMCID: PMC4314081
- DOI: 10.1371/journal.pone.0116839
Inhibitor of DNA Binding 4 (ID4) is highly expressed in human melanoma tissues and may function to restrict normal differentiation of melanoma cells
Abstract
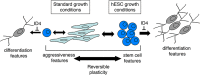
Melanoma tissues and cell lines are heterogeneous, and include cells with invasive, proliferative, stem cell-like, and differentiated properties. Such heterogeneity likely contributes to the aggressiveness of the disease and resistance to therapy. One model suggests that heterogeneity arises from rare cancer stem cells (CSCs) that produce distinct cancer cell lineages. Another model suggests that heterogeneity arises through reversible cellular plasticity, or phenotype-switching. Recent work indicates that phenotype-switching may include the ability of cancer cells to dedifferentiate to a stem cell-like state. We set out to investigate the phenotype-switching capabilities of melanoma cells, and used unbiased methods to identify genes that may control such switching. We developed a system to reversibly synchronize melanoma cells between 2D-monolayer and 3D-stem cell-like growth states. Melanoma cells maintained in the stem cell-like state showed a striking upregulation of a gene set related to development and neural stem cell biology, which included SRY-box 2 (SOX2) and Inhibitor of DNA Binding 4 (ID4). A gene set related to cancer cell motility and invasiveness was concomitantly downregulated. Intense and pervasive ID4 protein expression was detected in human melanoma tissue samples, suggesting disease relevance for this protein. SiRNA knockdown of ID4 inhibited switching from monolayer to 3D-stem cell-like growth, and instead promoted switching to a highly differentiated, neuronal-like morphology. We suggest that ID4 is upregulated in melanoma as part of a stem cell-like program that facilitates further adaptive plasticity. ID4 may contribute to disease by preventing stem cell-like melanoma cells from progressing to a normal differentiated state. This interpretation is guided by the known role of ID4 as a differentiation inhibitor during normal development. The melanoma stem cell-like state may be protected by factors such as ID4, thereby potentially identifying a new therapeutic vulnerability to drive differentiation to the normal cell phenotype.
Conflict of interest statement
Figures





Similar articles
-
ID4 imparts chemoresistance and cancer stemness to glioma cells by derepressing miR-9*-mediated suppression of SOX2.Cancer Res. 2011 May 1;71(9):3410-21. doi: 10.1158/0008-5472.CAN-10-3340. Cancer Res. 2011. PMID: 21531766
-
Regulation of glioblastoma multiforme stem-like cells by inhibitor of DNA binding proteins and oligodendroglial lineage-associated transcription factors.Cancer Sci. 2012 Jun;103(6):1028-37. doi: 10.1111/j.1349-7006.2012.02260.x. Epub 2012 Apr 4. Cancer Sci. 2012. PMID: 22380883 Free PMC article.
-
Melanoma spheroids grown under neural crest cell conditions are highly plastic migratory/invasive tumor cells endowed with immunomodulator function.PLoS One. 2011 Apr 15;6(4):e18784. doi: 10.1371/journal.pone.0018784. PLoS One. 2011. PMID: 21526207 Free PMC article.
-
Inhibitor of differentiation 4 (ID4): From development to cancer.Biochim Biophys Acta. 2015 Jan;1855(1):92-103. doi: 10.1016/j.bbcan.2014.12.002. Epub 2014 Dec 12. Biochim Biophys Acta. 2015. PMID: 25512197 Free PMC article. Review.
-
Cancer stem cells versus phenotype-switching in melanoma.Pigment Cell Melanoma Res. 2010 Dec;23(6):746-59. doi: 10.1111/j.1755-148X.2010.00757.x. Epub 2010 Aug 20. Pigment Cell Melanoma Res. 2010. PMID: 20726948 Review.
Cited by
-
Therapeutic implications of cellular and molecular biology of cancer stem cells in melanoma.Mol Cancer. 2017 Jan 30;16(1):7. doi: 10.1186/s12943-016-0578-3. Mol Cancer. 2017. PMID: 28137308 Free PMC article. Review.
-
Neural stem cells deriving from chick embryonic hindbrain recapitulate hindbrain development in culture.Sci Rep. 2018 Sep 17;8(1):13920. doi: 10.1038/s41598-018-32203-w. Sci Rep. 2018. PMID: 30224755 Free PMC article.
-
Id4 promotes cell proliferation in hepatocellular carcinoma.Chin J Cancer. 2017 Feb 1;36(1):19. doi: 10.1186/s40880-017-0186-7. Chin J Cancer. 2017. PMID: 28143562 Free PMC article.
-
ID1 and ID4 Are Biomarkers of Tumor Aggressiveness and Poor Outcome in Immunophenotypes of Breast Cancer.Cancers (Basel). 2021 Jan 27;13(3):492. doi: 10.3390/cancers13030492. Cancers (Basel). 2021. PMID: 33514024 Free PMC article.
-
New susceptibility loci for cutaneous melanoma risk and progression revealed using a porcine model.Oncotarget. 2018 Jun 12;9(45):27682-27697. doi: 10.18632/oncotarget.25455. eCollection 2018 Jun 12. Oncotarget. 2018. PMID: 29963229 Free PMC article.
References
-
- Miller AJ, Mihm MC Jr (2006) Melanoma. N Engl J Med 355: 51–65. - PubMed
-
- Sullivan RJ, Flaherty KT (2013) Resistance to BRAF-targeted therapy in melanoma. Eur J Cancer 49: 1297–1304. - PubMed
-
- Ohsie SJ, Sarantopoulos GP, Cochran AJ, Binder SW (2008) Immunohistochemical characteristics of melanoma. J Cutan Pathol 35: 433–444. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

