Structured observations reveal slow HIV-1 CTL escape
- PMID: 25642847
- PMCID: PMC4333731
- DOI: 10.1371/journal.pgen.1004914
Structured observations reveal slow HIV-1 CTL escape
Abstract
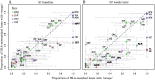
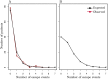
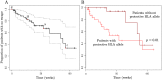
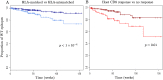
The existence of viral variants that escape from the selection pressures imposed by cytotoxic T-lymphocytes (CTLs) in HIV-1 infection is well documented, but it is unclear when they arise, with reported measures of the time to escape in individuals ranging from days to years. A study of participants enrolled in the SPARTAC (Short Pulse Anti-Retroviral Therapy at HIV Seroconversion) clinical trial allowed direct observation of the evolution of CTL escape variants in 125 adults with primary HIV-1 infection observed for up to three years. Patient HLA-type, longitudinal CD8+ T-cell responses measured by IFN-γ ELISpot and longitudinal HIV-1 gag, pol, and nef sequence data were used to study the timing and prevalence of CTL escape in the participants whilst untreated. Results showed that sequence variation within CTL epitopes at the first time point (within six months of the estimated date of seroconversion) was consistent with most mutations being transmitted in the infecting viral strain rather than with escape arising within the first few weeks of infection. Escape arose throughout the first three years of infection, but slowly and steadily. Approximately one third of patients did not drive any new escape in an HLA-restricted epitope in just under two years. Patients driving several escape mutations during these two years were rare and the median and modal numbers of new escape events in each patient were one and zero respectively. Survival analysis of time to escape found that possession of a protective HLA type significantly reduced time to first escape in a patient (p = 0.01), and epitopes escaped faster in the face of a measurable CD8+ ELISpot response (p = 0.001). However, even in an HLA matched host who mounted a measurable, specific, CD8+ response the average time before the targeted epitope evolved an escape mutation was longer than two years.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

