Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination
- PMID: 25642960
- PMCID: PMC4718306
- DOI: 10.1038/nature14157
Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination
Abstract
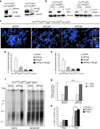
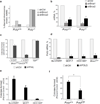
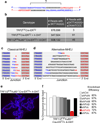
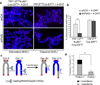
The alternative non-homologous end-joining (NHEJ) machinery facilitates several genomic rearrangements, some of which can lead to cellular transformation. This error-prone repair pathway is triggered upon telomere de-protection to promote the formation of deleterious chromosome end-to-end fusions. Using next-generation sequencing technology, here we show that repair by alternative NHEJ yields non-TTAGGG nucleotide insertions at fusion breakpoints of dysfunctional telomeres. Investigating the enzymatic activity responsible for the random insertions enabled us to identify polymerase theta (Polθ; encoded by Polq in mice) as a crucial alternative NHEJ factor in mammalian cells. Polq inhibition suppresses alternative NHEJ at dysfunctional telomeres, and hinders chromosomal translocations at non-telomeric loci. In addition, we found that loss of Polq in mice results in increased rates of homology-directed repair, evident by recombination of dysfunctional telomeres and accumulation of RAD51 at double-stranded breaks. Lastly, we show that depletion of Polθ has a synergistic effect on cell survival in the absence of BRCA genes, suggesting that the inhibition of this mutagenic polymerase represents a valid therapeutic avenue for tumours carrying mutations in homology-directed repair genes.
Figures
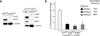
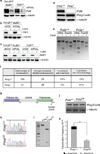


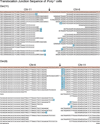

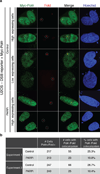


Comment in
-
DNA repair: Familiar ends with alternative endings.Nature. 2015 Feb 12;518(7538):174-6. doi: 10.1038/nature14200. Epub 2015 Feb 4. Nature. 2015. PMID: 25642961 No abstract available.
-
Targeted therapies: DNA polymerase θ-a new target for synthetic lethality?Nat Rev Clin Oncol. 2015 Mar;12(3):125. doi: 10.1038/nrclinonc.2015.23. Epub 2015 Feb 17. Nat Rev Clin Oncol. 2015. PMID: 25687907 No abstract available.
References
-
- van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

