Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair
- PMID: 25642963
- PMCID: PMC4415602
- DOI: 10.1038/nature14184
Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair
Abstract
Large-scale genomic studies have shown that half of epithelial ovarian cancers (EOCs) have alterations in genes regulating homologous recombination (HR) repair. Loss of HR accounts for the genomic instability of EOCs and for their cellular hyper-dependence on alternative poly-ADP ribose polymerase (PARP)-mediated DNA repair mechanisms. Previous studies have implicated the DNA polymerase θ (Polθ also known as POLQ, encoded by POLQ) in a pathway required for the repair of DNA double-strand breaks, referred to as the error-prone microhomology-mediated end-joining (MMEJ) pathway. Whether Polθ interacts with canonical DNA repair pathways to prevent genomic instability remains unknown. Here we report an inverse correlation between HR activity and Polθ expression in EOCs. Knockdown of Polθ in HR-proficient cells upregulates HR activity and RAD51 nucleofilament assembly, while knockdown of Polθ in HR-deficient EOCs enhances cell death. Consistent with these results, genetic inactivation of an HR gene (Fancd2) and Polq in mice results in embryonic lethality. Moreover, Polθ contains RAD51 binding motifs and it blocks RAD51-mediated recombination. Our results reveal a synthetic lethal relationship between the HR pathway and Polθ-mediated repair in EOCs, and identify Polθ as a novel druggable target for cancer therapy.
Figures





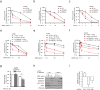


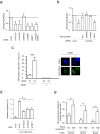


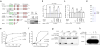


Comment in
-
DNA repair: Familiar ends with alternative endings.Nature. 2015 Feb 12;518(7538):174-6. doi: 10.1038/nature14200. Epub 2015 Feb 4. Nature. 2015. PMID: 25642961 No abstract available.
-
Targeted therapies: DNA polymerase θ-a new target for synthetic lethality?Nat Rev Clin Oncol. 2015 Mar;12(3):125. doi: 10.1038/nrclinonc.2015.23. Epub 2015 Feb 17. Nat Rev Clin Oncol. 2015. PMID: 25687907 No abstract available.
References
Additional References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

