A ranking method for the concurrent learning of compounds with various activity profiles
- PMID: 25643067
- PMCID: PMC4306736
- DOI: 10.1186/s13321-014-0050-6
A ranking method for the concurrent learning of compounds with various activity profiles
Abstract
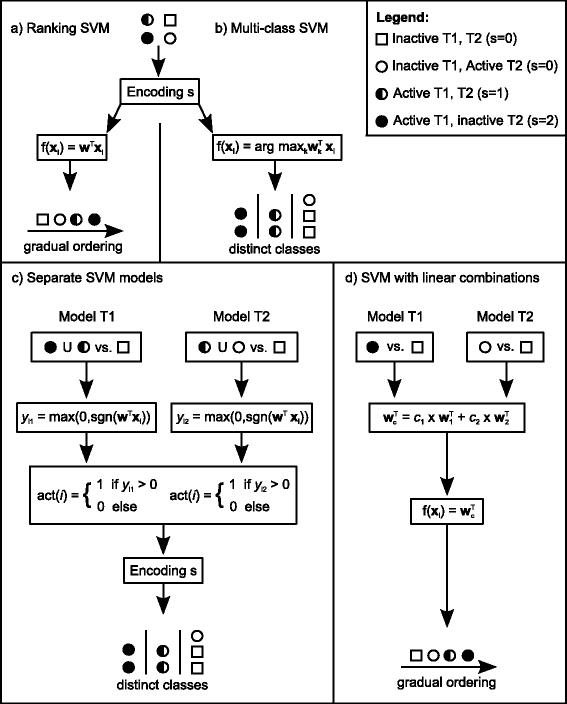
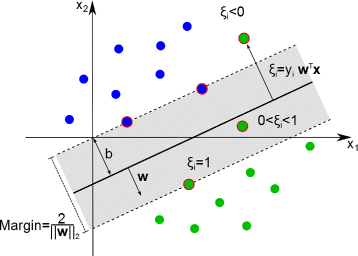
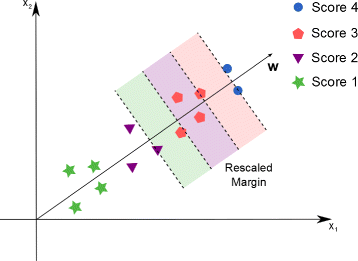
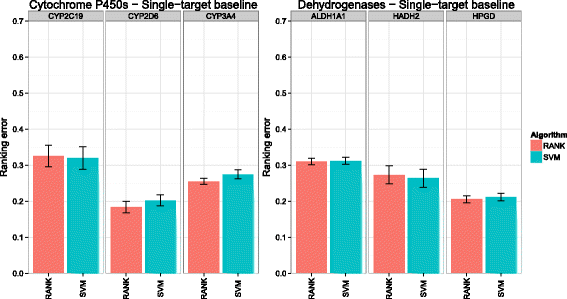
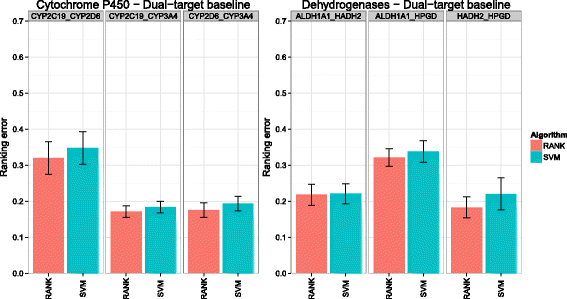
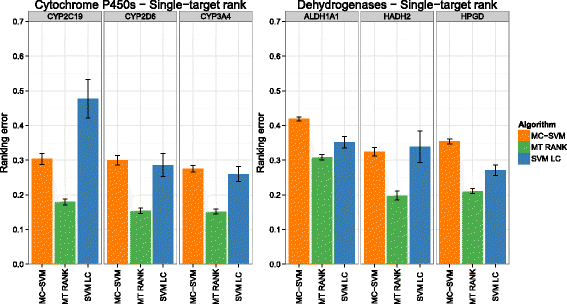
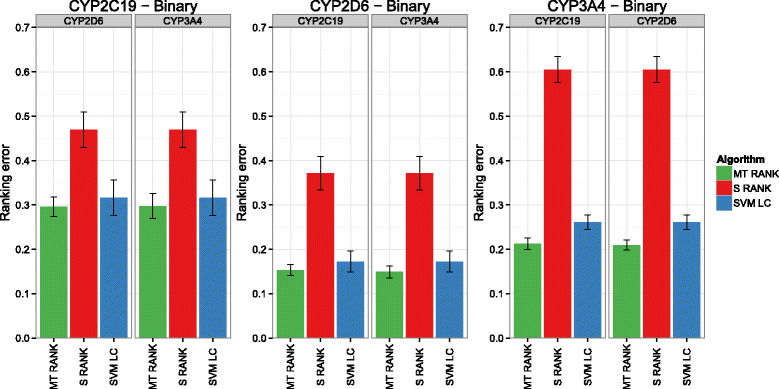
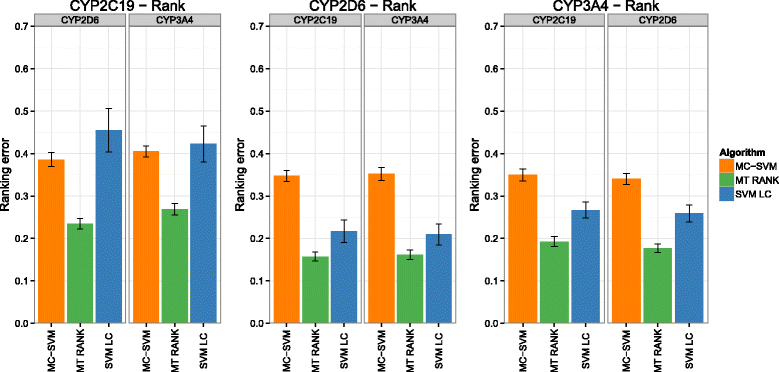
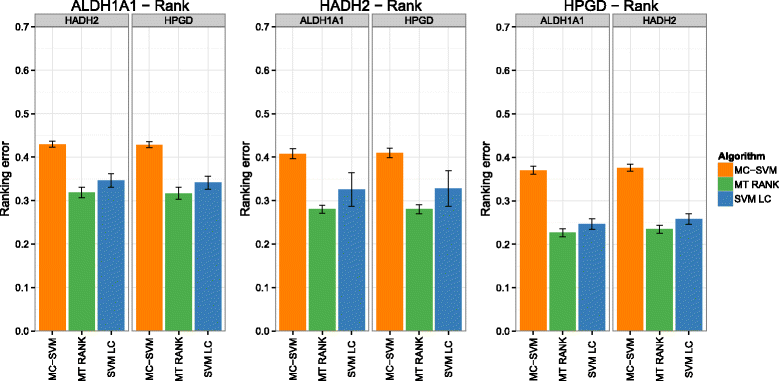
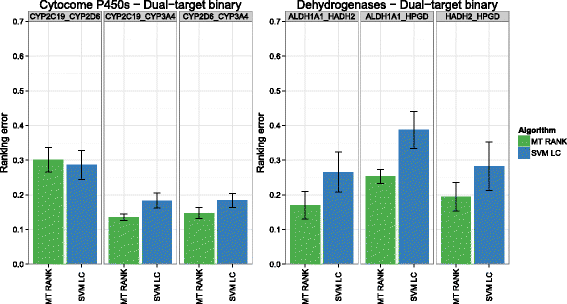
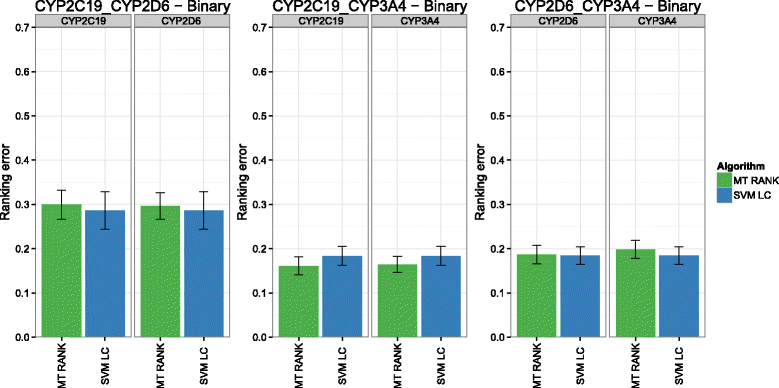
Background: In this study, we present a SVM-based ranking algorithm for the concurrent learning of compounds with different activity profiles and their varying prioritization. To this end, a specific labeling of each compound was elaborated in order to infer virtual screening models against multiple targets. We compared the method with several state-of-the-art SVM classification techniques that are capable of inferring multi-target screening models on three chemical data sets (cytochrome P450s, dehydrogenases, and a trypsin-like protease data set) containing three different biological targets each.
Results: The experiments show that ranking-based algorithms show an increased performance for single- and multi-target virtual screening. Moreover, compounds that do not completely fulfill the desired activity profile are still ranked higher than decoys or compounds with an entirely undesired profile, compared to other multi-target SVM methods.
Conclusions: SVM-based ranking methods constitute a valuable approach for virtual screening in multi-target drug design. The utilization of such methods is most helpful when dealing with compounds with various activity profiles and the finding of many ligands with an already perfectly matching activity profile is not to be expected.
Keywords: Machine learning; Multi-target; Ranking; Support vector machine; Virtual screening.
Figures









Similar articles
-
Evolution of Support Vector Machine and Regression Modeling in Chemoinformatics and Drug Discovery.J Comput Aided Mol Des. 2022 May;36(5):355-362. doi: 10.1007/s10822-022-00442-9. Epub 2022 Mar 19. J Comput Aided Mol Des. 2022. PMID: 35304657 Free PMC article.
-
Determination of Meta-Parameters for Support Vector Machine Linear Combinations.Mol Inform. 2015 Feb;34(2-3):127-33. doi: 10.1002/minf.201400163. Epub 2015 Feb 17. Mol Inform. 2015. PMID: 27490035
-
Ligand based virtual screening using SVM on GPU.Comput Biol Chem. 2019 Dec;83:107143. doi: 10.1016/j.compbiolchem.2019.107143. Epub 2019 Nov 10. Comput Biol Chem. 2019. PMID: 31743833
-
Drug Design for CNS Diseases: Polypharmacological Profiling of Compounds Using Cheminformatic, 3D-QSAR and Virtual Screening Methodologies.Front Neurosci. 2016 Jun 10;10:265. doi: 10.3389/fnins.2016.00265. eCollection 2016. Front Neurosci. 2016. PMID: 27375423 Free PMC article. Review.
-
A review of image analysis and machine learning techniques for automated cervical cancer screening from pap-smear images.Comput Methods Programs Biomed. 2018 Oct;164:15-22. doi: 10.1016/j.cmpb.2018.05.034. Epub 2018 Jun 26. Comput Methods Programs Biomed. 2018. PMID: 30195423 Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

