DNA interstrand cross-link repair requires replication-fork convergence
- PMID: 25643322
- PMCID: PMC4351167
- DOI: 10.1038/nsmb.2956
DNA interstrand cross-link repair requires replication-fork convergence
Abstract
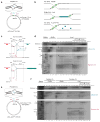
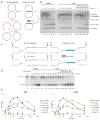
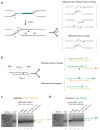
DNA interstrand cross-links (ICLs) prevent strand separation during DNA replication and transcription and therefore are extremely cytotoxic. In metazoans, a major pathway of ICL repair is coupled to DNA replication, and it requires the Fanconi anemia pathway. In most current models, collision of a single DNA replication fork with an ICL is sufficient to initiate repair. In contrast, we show here that in Xenopus egg extracts two DNA replication forks must converge on an ICL to trigger repair. When only one fork reaches the ICL, the replicative CMG helicase fails to unload from the stalled fork, and repair is blocked. Arrival of a second fork, even when substantially delayed, rescues repair. We conclude that ICL repair requires a replication-induced X-shaped DNA structure surrounding the lesion, and we speculate on how this requirement helps maintain genomic stability in S phase.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

