Amoebal endosymbiont Parachlamydia acanthamoebae Bn9 can grow in immortal human epithelial HEp-2 cells at low temperature; an in vitro model system to study chlamydial evolution
- PMID: 25643359
- PMCID: PMC4314085
- DOI: 10.1371/journal.pone.0116486
Amoebal endosymbiont Parachlamydia acanthamoebae Bn9 can grow in immortal human epithelial HEp-2 cells at low temperature; an in vitro model system to study chlamydial evolution
Abstract
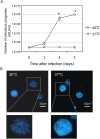
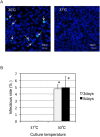
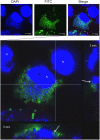
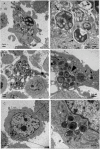
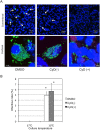
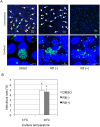
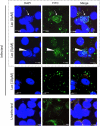
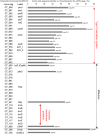
Ancient chlamydiae diverged into pathogenic and environmental chlamydiae 0.7-1.4 billion years ago. However, how pathogenic chlamydiae adapted to mammalian cells that provide a stable niche at approximately 37 °C, remains unknown, although environmental chlamydiae have evolved as endosymbionts of lower eukaryotes in harsh niches of relatively low temperatures. Hence, we assessed whether an environmental chlamydia, Parachlamydia Bn9, could grow in human HEp-2 cells at a low culture temperature of 30 °C. The assessment of inclusion formation by quantitative RT-PCR revealed that the numbers of bacterial inclusion bodies and the transcription level of 16SrRNA significantly increased after culture at 30 °C compared to at 37 °C. Confocal microscopy showed that the bacteria were located close to HEp-2 nuclei and were actively replicative. Transmission electron microscopy also revealed replicating bacteria consisting of reticular bodies, but with a few elementary bodies. Cytochalasin D and rifampicin inhibited inclusion formation. Lactacystin slightly inhibited bacterial inclusion formation. KEGG analysis using a draft genome sequence of the bacteria revealed that it possesses metabolic pathways almost identical to those of pathogenic chlamydia. Interestingly, comparative genomic analysis with pathogenic chlamydia revealed that the Parachlamydia similarly possess the genes encoding Type III secretion system, but lacking genes encoding inclusion membrane proteins (IncA to G) required for inclusion maturation. Taken together, we conclude that ancient chlamydiae had the potential to grow in human cells, but overcoming the thermal gap was a critical event for chlamydial adaptation to human cells.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

