ID helix-loop-helix proteins as determinants of cell survival in B-cell chronic lymphocytic leukemia cells in vitro
- PMID: 25644253
- PMCID: PMC4320821
- DOI: 10.1186/s12943-014-0286-9
ID helix-loop-helix proteins as determinants of cell survival in B-cell chronic lymphocytic leukemia cells in vitro
Abstract
Background: Members of the inhibitor of DNA-binding (ID) family of helix-loop-helix proteins have been causally implicated in the pathogenesis of several types of B-cell lineage malignancy, either on the basis of mutation or by altered expression. B-cell chronic lymphocytic leukemia encompasses a heterogeneous group of disorders and is the commonest leukaemia type in the Western world. In this study, we have investigated the pathobiological functions of the ID2 and ID3 proteins in this disease with an emphasis on their role in regulating leukemic cell death/survival.
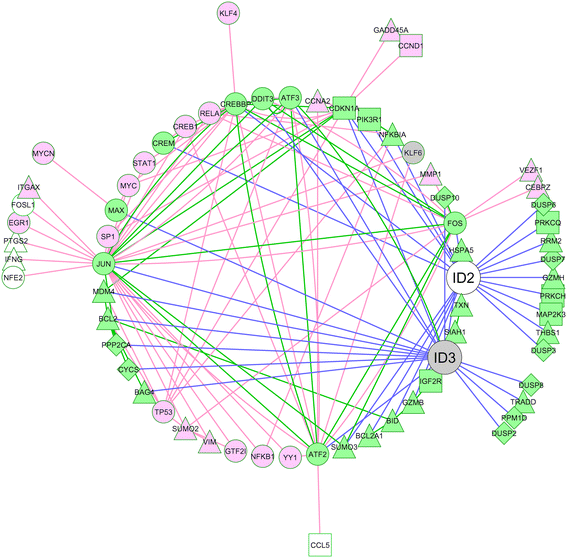
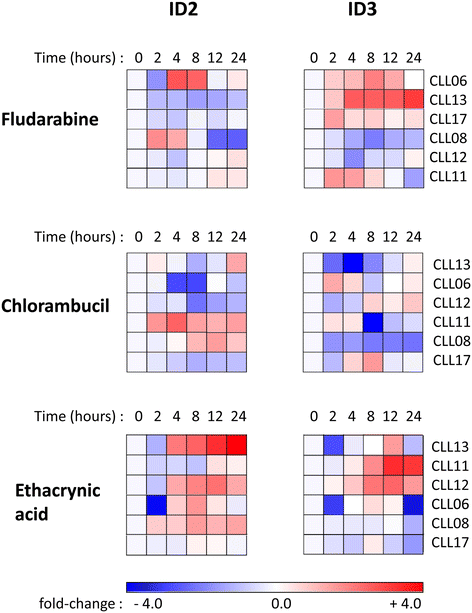
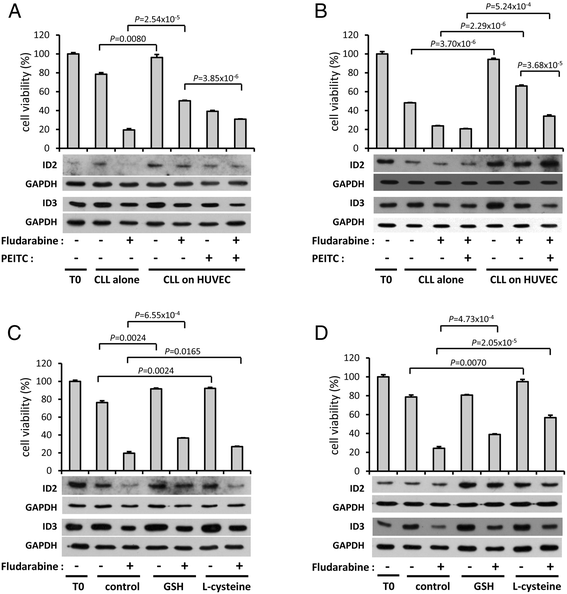
Methods: Bioinformatics analysis of microarray gene expression data was used to investigate expression of ID2/ID3 in leukemic versus normal B cells, their association with clinical course of disease and molecular sub-type and to reconstruct a gene regulatory network using the 'maximum information coefficient' (MIC) for target gene inference. In vitro cultured primary leukemia cells, either in isolation or co-cultured with accessory vascular endothelial cells, were used to investigate ID2/ID3 protein expression by western blotting and to assess the cytotoxic response of different drugs (fludarabine, chlorambucil, ethacrynic acid) by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. ID2/ID3 protein levels in primary leukemia cells and in MEC1 cells were manipulated by transduction with siRNA reagents.
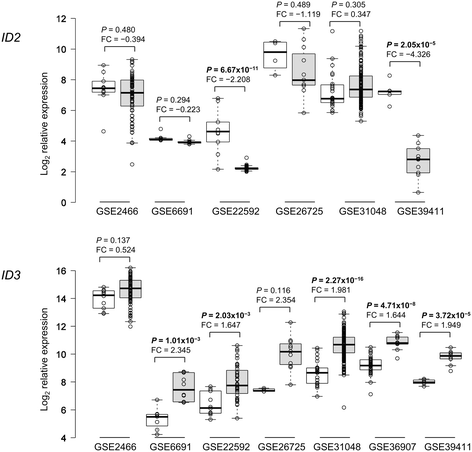
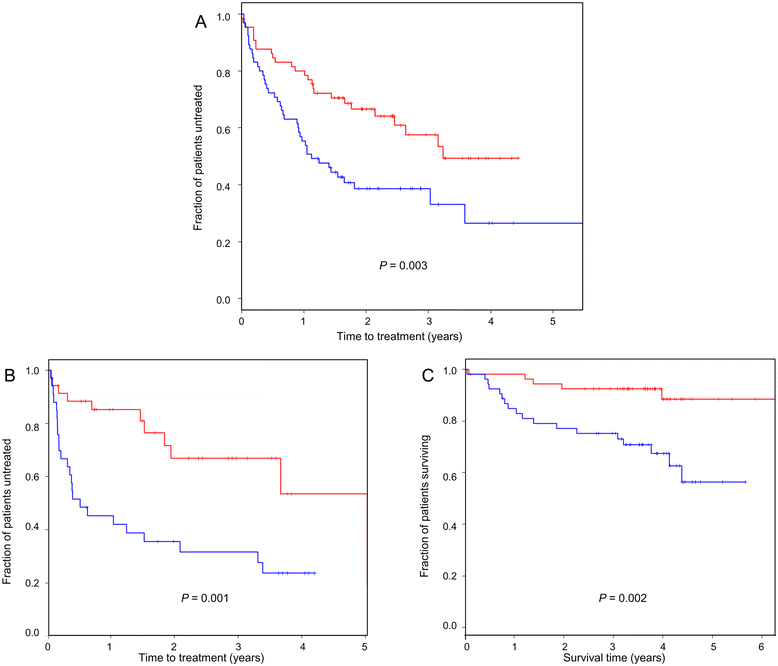
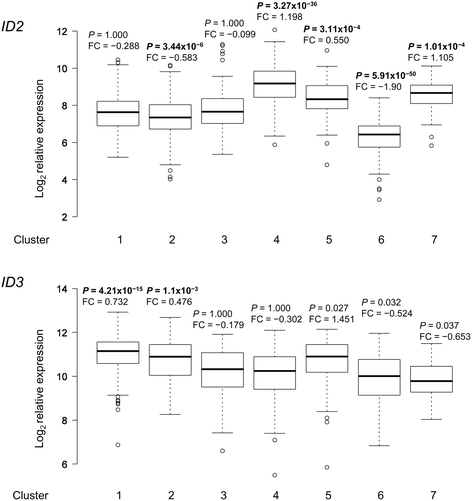
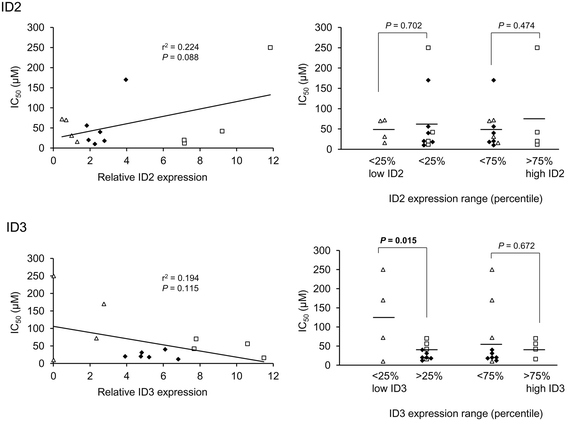
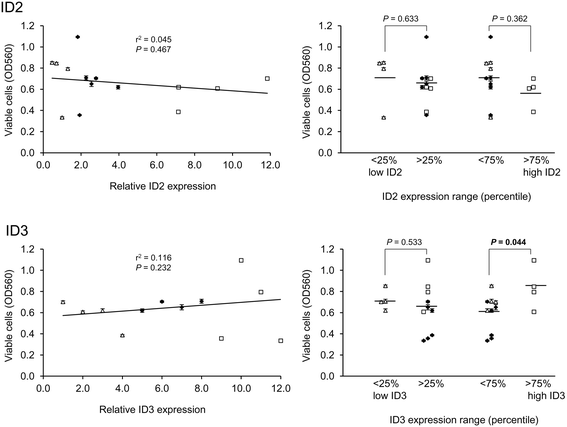
Results: Datamining showed that the expression profiles of ID2 and ID3 are associated with distinct pathobiological features of disease and implicated both genes in regulating cell death/survival by targeting multiple non-overlapping sets of apoptosis effecter genes. Consistent with microarray data, the overall pattern of ID2/ID3 protein expression in relation to cell death/survival responses of primary leukemia cells was suggestive of a pro-survival function for both ID proteins. This was confirmed by siRNA knock-down experiments in MEC1 cells and in primary leukemia cells, but with variability in the dependence of leukemic cells from different patients on ID protein expression for cell survival. Vascular endothelial cells rescued leukemia cells from spontaneous and cytotoxic drug-induced cell death at least in part, via an ID protein-coupled redox-dependent mechanism.
Conclusions: Our study provides evidence for a pro-survival function of the ID2/ID3 proteins in chronic lymphocytic leukemia cells and also highlights these proteins as potential determinants of the pathobiology of this disorder.
Figures




References
-
- Norton JD. ID helix-loop-helix proteins in cell growth, differentiation and tumourigenesis. J Cell Sci. 2000;113:3897–3905. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

