Molecular insights into the binding of coenzyme F420 to the conserved protein Rv1155 from Mycobacterium tuberculosis
- PMID: 25644473
- PMCID: PMC4420522
- DOI: 10.1002/pro.2645
Molecular insights into the binding of coenzyme F420 to the conserved protein Rv1155 from Mycobacterium tuberculosis
Abstract
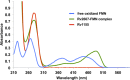
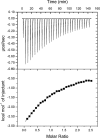
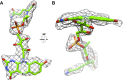
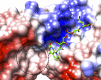
Coenzyme F420 is a deazaflavin hydride carrier with a lower reduction potential than most flavins. In Mycobacterium tuberculosis (Mtb), F420 plays an important role in activating PA-824, an antituberculosis drug currently used in clinical trials. Although F420 is important to Mtb redox metabolism, little is known about the enzymes that bind F420 and the reactions that they catalyze. We have identified a novel F420 -binding protein, Rv1155, which is annotated in the Mtb genome sequence as a putative flavin mononucleotide (FMN)-binding protein. Using biophysical techniques, we have demonstrated that instead of binding FMN or other flavins, Rv1155 binds coenzyme F420 . The crystal structure of the complex of Rv1155 and F420 reveals one F420 molecule bound to each monomer of the Rv1155 dimer. Structural, biophysical, and bioinformatic analyses of the Rv1155-F420 complex provide clues about its role in the bacterium.
Keywords: Mycobacterium tuberculosis; Rv1155; coenzyme F420; conserved hypothetical protein.
© 2015 The Protein Society.
Figures


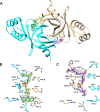
References
-
- Koul A, Arnoult E, Lounis N, Guillemont J, Andries K. The challenge of new drug discovery for tuberculosis. Nature. 2011;469:483–490. - PubMed
-
- Raviglione M, Marais B, Floyd K, Lonnroth K, Getahun H, Migliori GB, Harries AD, Nunn P, Lienhardt C, Graham S, Chakaya J, Weyer K, Cole S, Kaufmann SH, Zumla A. Scaling up interventions to achieve global tuberculosis control: progress and new developments. Lancet. 2012;379:1902–1913. - PubMed
-
- Barry CE, III, Boshoff HI, Dowd CS. Prospects for clinical introduction of nitroimidazole antibiotics for the treatment of tuberculosis. Curr Pharm Des. 2004;10:3239–3262. - PubMed
-
- Bair TB, Isabelle DW, Daniels L. Structures of coenzyme F420 in Mycobacterium species. Arch Microbiol. 2001;176:37–43. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

