Whole-epigenome analysis in multiple myeloma reveals DNA hypermethylation of B cell-specific enhancers
- PMID: 25644835
- PMCID: PMC4381520
- DOI: 10.1101/gr.180240.114
Whole-epigenome analysis in multiple myeloma reveals DNA hypermethylation of B cell-specific enhancers
Abstract
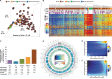
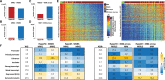
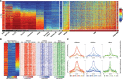
While analyzing the DNA methylome of multiple myeloma (MM), a plasma cell neoplasm, by whole-genome bisulfite sequencing and high-density arrays, we observed a highly heterogeneous pattern globally characterized by regional DNA hypermethylation embedded in extensive hypomethylation. In contrast to the widely reported DNA hypermethylation of promoter-associated CpG islands (CGIs) in cancer, hypermethylated sites in MM, as opposed to normal plasma cells, were located outside CpG islands and were unexpectedly associated with intronic enhancer regions defined in normal B cells and plasma cells. Both RNA-seq and in vitro reporter assays indicated that enhancer hypermethylation is globally associated with down-regulation of its host genes. ChIP-seq and DNase-seq further revealed that DNA hypermethylation in these regions is related to enhancer decommissioning. Hypermethylated enhancer regions overlapped with binding sites of B cell-specific transcription factors (TFs) and the degree of enhancer methylation inversely correlated with expression levels of these TFs in MM. Furthermore, hypermethylated regions in MM were methylated in stem cells and gradually became demethylated during normal B-cell differentiation, suggesting that MM cells either reacquire epigenetic features of undifferentiated cells or maintain an epigenetic signature of a putative myeloma stem cell progenitor. Overall, we have identified DNA hypermethylation of developmentally regulated enhancers as a new type of epigenetic modification associated with the pathogenesis of MM.
© 2015 Agirre et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


References
-
- Bergsagel PL, Kuehl WM. 2005. Molecular pathogenesis and a consequent classification of multiple myeloma. J Clin Oncol 23: 6333–6338. - PubMed
-
- Berman BP, Weisenberger DJ, Aman JF, Hinoue T, Ramjan Z, Liu Y, Noushmehr H, Lange CP, van Dijk CM, Tollenaar RA, et al.2011. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet 44: 40–46. - PMC - PubMed
-
- Busche S, Ge B, Vidal R, Spinella JF, Saillour V, Richer C, Healy J, Chen SH, Droit A, Sinnett D, et al.2013. Integration of high-resolution methylome and transcriptome analyses to dissect epigenomic changes in childhood acute lymphoblastic leukemia. Cancer Res 73: 4323–4336. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
