ZNF143 provides sequence specificity to secure chromatin interactions at gene promoters
- PMID: 25645053
- PMCID: PMC4431651
- DOI: 10.1038/ncomms7186
ZNF143 provides sequence specificity to secure chromatin interactions at gene promoters
Erratum in
-
Publisher Correction: ZNF143 provides sequence specificity to secure chromatin interactions at gene promoters.Nat Commun. 2018 Apr 10;9:16194. doi: 10.1038/ncomms16194. Nat Commun. 2018. PMID: 29633758 Free PMC article.
Abstract
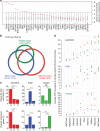
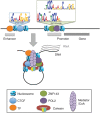
Chromatin interactions connect distal regulatory elements to target gene promoters guiding stimulus- and lineage-specific transcription. Few factors securing chromatin interactions have so far been identified. Here, by integrating chromatin interaction maps with the large collection of transcription factor-binding profiles provided by the ENCODE project, we demonstrate that the zinc-finger protein ZNF143 preferentially occupies anchors of chromatin interactions connecting promoters with distal regulatory elements. It binds directly to promoters and associates with lineage-specific chromatin interactions and gene expression. Silencing ZNF143 or modulating its DNA-binding affinity using single-nucleotide polymorphisms (SNPs) as a surrogate of site-directed mutagenesis reveals the sequence dependency of chromatin interactions at gene promoters. We also find that chromatin interactions alone do not regulate gene expression. Together, our results identify ZNF143 as a novel chromatin-looping factor that contributes to the architectural foundation of the genome by providing sequence specificity at promoters connected with distal regulatory elements.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

