Quantitative and logic modelling of molecular and gene networks
- PMID: 25645874
- PMCID: PMC4604653
- DOI: 10.1038/nrg3885
Quantitative and logic modelling of molecular and gene networks
Abstract
Behaviours of complex biomolecular systems are often irreducible to the elementary properties of their individual components. Explanatory and predictive mathematical models are therefore useful for fully understanding and precisely engineering cellular functions. The development and analyses of these models require their adaptation to the problems that need to be solved and the type and amount of available genetic or molecular data. Quantitative and logic modelling are among the main methods currently used to model molecular and gene networks. Each approach comes with inherent advantages and weaknesses. Recent developments show that hybrid approaches will become essential for further progress in synthetic biology and in the development of virtual organisms.
Figures

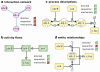

References
-
- Tindall MJ, Gaffney E. a, Maini PK, Armitage JP. Theoretical insights into bacterial chemotaxis. Wiley Interdiscip. Rev. Syst. Biol. Med. 2012;4:247–59. - PubMed
-
- Karr JR, et al. A whole-cell computational model predicts phenotype from genotype. Cell. 2012;150:389–401. - PMC - PubMed
-
Modular model of an entire Mycoplasma genitalium cell, including the expression of all genes, all metabolites and signalling pathways. The model is simulated with an hybrid approach, using stochastic simulations, ODEs and flux balance analysis.
-
- Schliess F, et al. Integrated metabolic spatial-temporal model for the prediction of ammonia detoxification during liver damage and regeneration. Hepatology. 2014:2–44. doi:10.1002/hep.27136. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

