Anti-tumoral effects of miR-3189-3p in glioblastoma
- PMID: 25645911
- PMCID: PMC4375465
- DOI: 10.1074/jbc.M114.633081
Anti-tumoral effects of miR-3189-3p in glioblastoma
Abstract
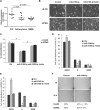
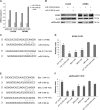
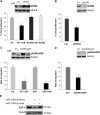
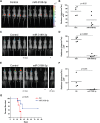
Glioblastoma is one of the most aggressive brain tumors. We have previously found up-regulation of growth differentiation factor 15 (GDF15) in glioblastoma cells treated with the anticancer agent fenofibrate. Sequence analysis of GDF15 revealed the presence of a microRNA, miR-3189, in the single intron. We then asked whether miR-3189 was expressed in clinical samples and whether it was functional in glioblastoma cells. We found that expression of miR-3189-3p was down-regulated in astrocytoma and glioblastoma clinical samples compared with control brain tissue. In vitro, the functionality of miR-3189-3p was tested by RNA-binding protein immunoprecipitation, and miR-3189-3p coimmunoprecipitated with Argonaute 2 together with two of its major predicted gene targets, the SF3B2 splicing factor and the guanine nucleotide exchange factor p63RhoGEF. Overexpression of miR-3189-3p resulted in a significant inhibition of cell proliferation and migration through direct targeting of SF3B2 and p63RhoGEF, respectively. Interestingly, miR-3189-3p levels were increased by treatment of glioblastoma cells with fenofibrate, a lipid-lowering drug with multiple anticancer activities. The attenuated expression of miR-3189-3p in clinical samples paralleled the elevated expression of SF3B2, which could contribute to the activation of SF3B2 growth-promoting pathways in these tumors. Finally, miR-3189-3p-mediated inhibition of tumor growth in vivo further supported the function of this microRNA as a tumor suppressor.
Keywords: Cell Migration; Cell Proliferation; Glioblastoma; MicroRNA (miRNA); Tumor Suppressor Gene.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



References
-
- Nagasawa D. T., Chow F., Yew A., Kim W., Cremer N., Yang I. (2012) Temozolomide and other potential agents for the treatment of glioblastoma multiforme. Neurosurg. Clin. N. Am. 23, 307–322, ix - PubMed
-
- Hegi M. E., Diserens A. C., Gorlia T., Hamou M. F., de Tribolet N., Weller M., Kros J. M., Hainfellner J. A., Mason W., Mariani L., Bromberg J. E., Hau P., Mirimanoff R. O., Cairncross J. G., Janzer R. C., Stupp R. (2005) MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 352, 997–1003 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

