SAMHD1's protein expression profile in humans
- PMID: 25646359
- PMCID: PMC7166976
- DOI: 10.1189/jlb.4HI0714-338RR
SAMHD1's protein expression profile in humans
Abstract
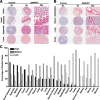
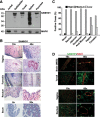
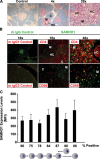
The deoxynucleoside triphosphate triphosphohydrolase and 3' → 5' exonuclease SAMHD1 restricts HIV-1 infection in noncycling hematopoietic cells in vitro, and SAMHD1 mutations are associated with AGS. Little is known about the in vivo expression and functional regulation of this cellular factor. Here, we first assessed the SAMHD1 protein expression profile on a microarray of 25 human tissues from >210 donors and in purified primary cell populations. In vivo, SAMHD1 was expressed in the majority of nucleated cells of hematopoietic origin, including tissue-resident macrophages, DCs, pDCs, all developmental stages of thymic T cells, monocytes, NK cells, as well as at lower levels in B cells. Of note, SAMHD1 was abundantly expressed in HIV target cells residing in the anogenital mucosa, providing a basis for its evaluation as a cellular factor that may impact the efficiency of HIV transmission. Next, we examined the effect of the activation status and proinflammatory cytokine treatment of cells on expression and phosphorylation of SAMHD1. Activated, HIV-susceptible CD4(+) T cells carried pSAMHD1(T592), whereas resting CD4(+) T cells and macrophages expressed the unphosphorylated protein with HIV-restrictive activity. Surprisingly, stimulation of these primary cells with IFN-α, IFN-γ, IL-4, IL-6, IL-12, IL-18, IL-27, or TNF-α affected neither SAMHD1 expression levels nor threonine 592 phosphorylation. Only IL-1β moderately down-regulated SAMHD1 in activated CD4(+) T cells. Taken together, this study establishes the first cross-sectional protein expression profile of SAMHD1 in human tissues and provides insight into its cell cycle-dependent phosphorylation and unresponsiveness to multiple proinflammatory cytokines.
Keywords: HIV; HSV; innate immunity; restriction factor.
© Society for Leukocyte Biology.
Figures



Comment in
-
Editorial: Passive aggressive avoidance of SAMHD1 restriction by HIV-1.J Leukoc Biol. 2015 Jul;98(1):1-3. doi: 10.1189/jlb.4CE0215-044R. J Leukoc Biol. 2015. PMID: 26130765 No abstract available.
References
-
- Goutieres, F. (2005) Aicardi‐Goutieres syndrome. Brain Dev. 27, 201–206. - PubMed
-
- Li, N. , Zhang, W. , Cao, X. (2000) Identification of human homologue of mouse IFN‐gamma induced protein from human dendritic cells. Immunol. Lett. 74, 221–224. - PubMed
-
- Rice, G.I. , Bond, J. , Asipu, A. , Brunette, R.L. , Manfield, I.W. , Carr, I.M. , Fuller, J.C. , Jackson, R.M. , Lamb, T. , Briggs, T.A. , Ali, M. , Gornall, H. , Couthard, L.R. , Aeby, A. , Attard‐Montalto, S.P. , Bertini, E. , Bodemer, C. , Brockmann, K. , Brueton, L.A. , Corry, P.C. , Desguerre, I. , Fazzi, E. , Cazorla, A.G. , Gener, B. , Hamel, B.C. , Heiberg, A. , Hunter, M. , van der Knaap, M.S. , Kumar, R. , Lagae, L. , Landrieu, P.G. , Lourenco, C.M. , Marom, D. , McDermott, M.F. , van der Merwe, W. , Orcesi, S. , Prendiville, J.S. , Rasmussen, M. , Shalev, S.A. , Soler, D.M. , Shinawi, M. , Spiegel, R. , Tan, T.Y. , Vanderver, A. , Wakeling, E.L. , Wassmer, E. , Whittaker, E. , Lebon, P. , Stetson, D.B. , Bonthron, D.T. , Crow, Y.J. (2009) Mutations involved in Aicardi‐Goutièeres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat. Genet. 41, 829–832. - PMC - PubMed
-
- Liao, W. , Bao, Z. , Cheng, C. , Mok, Y.K. , Wong, W.S. (2008) Dendritic cell‐derived interferon‐gamma‐induced protein mediates tumor necrosis factor‐alpha stimulation of human lung fibroblasts. Proteomics 8, 2640–2650. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

