Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients
- PMID: 25646427
- PMCID: PMC4372002
- DOI: 10.1073/pnas.1500076112
Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients
Abstract
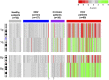
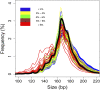
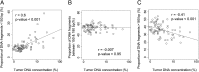
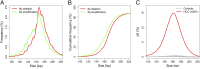
The analysis of tumor-derived circulating cell-free DNA opens up new possibilities for performing liquid biopsies for the assessment of solid tumors. Although its clinical potential has been increasingly recognized, many aspects of the biological characteristics of tumor-derived cell-free DNA remain unclear. With respect to the size profile of such plasma DNA molecules, a number of studies reported the finding of increased integrity of tumor-derived plasma DNA, whereas others found evidence to suggest that plasma DNA molecules released by tumors might be shorter. Here, we performed a detailed analysis of the size profiles of plasma DNA in 90 patients with hepatocellular carcinoma, 67 with chronic hepatitis B, 36 with hepatitis B-associated cirrhosis, and 32 healthy controls. We used massively parallel sequencing to achieve plasma DNA size measurement at single-base resolution and in a genome-wide manner. Tumor-derived plasma DNA molecules were further identified with the use of chromosome arm-level z-score analysis (CAZA), which facilitated the studying of their specific size profiles. We showed that populations of aberrantly short and long DNA molecules existed in the plasma of patients with hepatocellular carcinoma. The short ones preferentially carried the tumor-associated copy number aberrations. We further showed that there were elevated amounts of plasma mitochondrial DNA in the plasma of hepatocellular carcinoma patients. Such molecules were much shorter than the nuclear DNA in plasma. These results have improved our understanding of the size profile of tumor-derived circulating cell-free DNA and might further enhance our ability to use plasma DNA as a molecular diagnostic tool.
Keywords: circulating tumor DNA; liquid biopsy; massively parallel sequencing; mitochondrial DNA; tumor markers.
Conflict of interest statement
Conflict of interest statement: R.W.K.C. and Y.M.D.L. received research support from Sequenom, Inc. R.W.K.C. and Y.M.D.L. are consultants to Sequenom, Inc. R.W.K.C., K.C.A.C., and Y.M.D.L. hold equities in Sequenom, Inc. R.W.K.C., K.C.A.C., and Y.M.D.L. are founders of Xcelom. P.J., R.W.K.C., K.C.A.C., and Y.M.D.L. have filed patents/patent applications based on the data generated from this work.
Figures


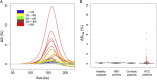

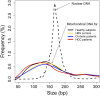
Comment in
-
Circulating tumor-derived DNA is shorter than somatic DNA in plasma.Proc Natl Acad Sci U S A. 2015 Mar 17;112(11):3178-9. doi: 10.1073/pnas.1501321112. Epub 2015 Mar 2. Proc Natl Acad Sci U S A. 2015. PMID: 25733911 Free PMC article. No abstract available.
References
-
- Jung K, Fleischhacker M, Rabien A. Cell-free DNA in the blood as a solid tumor biomarker—a critical appraisal of the literature. Clin Chim Acta. 2010;411(21-22):1611–1624. - PubMed
-
- Chan KCA. Scanning for cancer genomic changes in plasma: Toward an era of personalized blood-based tumor markers. Clin Chem. 2013;59(11):1553–1555. - PubMed
-
- Bidard FC, Weigelt B, Reis-Filho JS. Going with the flow: From circulating tumor cells to DNA. Sci Transl Med. 2013;5(207):07ps14. - PubMed
-
- Beck J, Urnovitz HB, Mitchell WM, Schütz E. Next generation sequencing of serum circulating nucleic acids from patients with invasive ductal breast cancer reveals differences to healthy and nonmalignant controls. Mol Cancer Res. 2010;8(3):335–342. - PubMed
-
- Dawson SJ, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368(13):1199–1209. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

