Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development
- PMID: 25646447
- PMCID: PMC4343106
- DOI: 10.1073/pnas.1500365112
Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development
Abstract
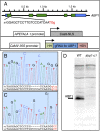
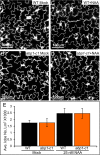
Auxin binding protein 1 (ABP1) has been studied for decades. It has been suggested that ABP1 functions as an auxin receptor and has an essential role in many developmental processes. Here we present our unexpected findings that ABP1 is neither required for auxin signaling nor necessary for plant development under normal growth conditions. We used our ribozyme-based CRISPR technology to generate an Arabidopsis abp1 mutant that contains a 5-bp deletion in the first exon of ABP1, which resulted in a frameshift and introduction of early stop codons. We also identified a T-DNA insertion abp1 allele that harbors a T-DNA insertion located 27 bp downstream of the ATG start codon in the first exon. We show that the two new abp1 mutants are null alleles. Surprisingly, our new abp1 mutant plants do not display any obvious developmental defects. In fact, the mutant plants are indistinguishable from wild-type plants at every developmental stage analyzed. Furthermore, the abp1 plants are not resistant to exogenous auxin. At the molecular level, we find that the induction of known auxin-regulated genes is similar in both wild-type and abp1 plants in response to auxin treatments. We conclude that ABP1 is not a key component in auxin signaling or Arabidopsis development.
Keywords: ABP1; CRISPR; auxin; plant development; receptor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Comment in
-
Auxin Binding Protein 1: A Red Herring After All?Mol Plant. 2015 Aug;8(8):1131-4. doi: 10.1016/j.molp.2015.04.010. Epub 2015 Apr 25. Mol Plant. 2015. PMID: 25917757 No abstract available.
References
-
- Jones A. Auxin-binding proteins. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:393–420.
-
- Chen X, et al. ABP1 and ROP6 GTPase signaling regulate clathrin-mediated endocytosis in Arabidopsis roots. Curr Biol. 2012;22(14):1326–1332. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

