Overlapping hotspots in CDRs are critical sites for V region diversification
- PMID: 25646473
- PMCID: PMC4343087
- DOI: 10.1073/pnas.1500788112
Overlapping hotspots in CDRs are critical sites for V region diversification
Abstract
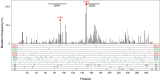
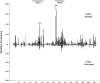
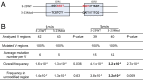
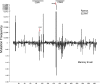
Activation-induced deaminase (AID) mediates the somatic hypermutation (SHM) of Ig variable (V) regions that is required for the affinity maturation of the antibody response. An intensive analysis of a published database of somatic hypermutations that arose in the IGHV3-23*01 human V region expressed in vivo by human memory B cells revealed that the focus of mutations in complementary determining region (CDR)1 and CDR2 coincided with a combination of overlapping AGCT hotspots, the absence of AID cold spots, and an abundance of polymerase eta hotspots. If the overlapping hotspots in the CDR1 or CDR2 did not undergo mutation, the frequency of mutations throughout the V region was reduced. To model this result, we examined the mutation of the human IGHV3-23*01 biochemically and in the endogenous heavy chain locus of Ramos B cells. Deep sequencing revealed that IGHV3-23*01 in Ramos cells accumulates AID-induced mutations primarily in the AGCT in CDR2, which was also the most frequent site of mutation in vivo. Replacing the overlapping hotspots in CDR1 and CDR2 with neutral or cold motifs resulted in a reduction in mutations within the modified motifs and, to some degree, throughout the V region. In addition, some of the overlapping hotspots in the CDRs were at sites in which replacement mutations could change the structure of the CDR loops. Our analysis suggests that the local sequence environment of the V region, and especially of the CDR1 and CDR2, is highly evolved to recruit mutations to key residues in the CDRs of the IgV region.
Keywords: AID; IGHV3-23; complementarity-determining regions; cytosine deamination; somatic hypermutation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Rogozin IB, Diaz M. Cutting edge: DGYW/WRCH is a better predictor of mutability at G:C bases in Ig hypermutation than the widely accepted RGYW/WRCY motif and probably reflects a two-step activation-induced cytidine deaminase-triggered process. J Immunol. 2004;172(6):3382–3384. - PubMed
-
- Zhang W, et al. Clonal instability of V region hypermutation in the Ramos Burkitt’s lymphoma cell line. Int Immunol. 2001;13(9):1175–1184. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 5R01CA072649/CA/NCI NIH HHS/United States
- 1R01GM111741-01A1/GM/NIGMS NIH HHS/United States
- R01 ES013192/ES/NIEHS NIH HHS/United States
- R01CA164468/CA/NCI NIH HHS/United States
- R01DA033788/DA/NIDA NIH HHS/United States
- 9R01AI112335/AI/NIAID NIH HHS/United States
- R01 CA164468/CA/NCI NIH HHS/United States
- R01 DA033788/DA/NIDA NIH HHS/United States
- R01 GM111741/GM/NIGMS NIH HHS/United States
- R56 ES013192/ES/NIEHS NIH HHS/United States
- R01 AI112335/AI/NIAID NIH HHS/United States
- R01 CA072649/CA/NCI NIH HHS/United States
- P30 CA013330/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

