Identification of destabilizing and stabilizing mutations of Ste2p, a G protein-coupled receptor in Saccharomyces cerevisiae
- PMID: 25647246
- PMCID: PMC4405178
- DOI: 10.1021/bi501314t
Identification of destabilizing and stabilizing mutations of Ste2p, a G protein-coupled receptor in Saccharomyces cerevisiae
Abstract
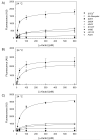
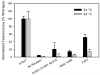
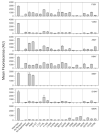
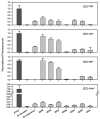
The isolation of mutations affecting the stabilities of transmembrane proteins is useful for enhancing the suitability of proteins for structural characterization and identification of determinants of membrane protein stability. We have pursued a strategy for the identification of stabilized variants of the yeast α-factor receptor Ste2p. Because it was not possible to screen directly for mutations providing thermal stabilization, we first isolated a battery of destabilized temperature-sensitive variants, based on loss of signaling function and decreased levels of binding of the fluorescent ligand, and then screened for intragenic second-site suppressors of these phenotypes. The initial screens recovered singly and multiply substituted mutations conferring temperature sensitivity throughout the predicted transmembrane helices of the receptor. All of the singly substituted variants exhibit decreases in cell-surface expression. We then screened randomly mutagenized libraries of clones expressing temperature-sensitive variants for second-site suppressors that restore elevated levels of binding sites for fluorescent ligand. To determine whether any of these were global suppressors, and thus likely stabilizing mutations, they were combined with different temperature-sensitive mutations. Eight of the suppressors exhibited the ability to reverse the defect in ligand binding of multiple temperature-sensitive mutations. Combining certain suppressors into a single allele resulted in levels of suppression greater than that seen with either suppressor alone. Solubilized receptors containing suppressor mutations in the absence of temperature-sensitive mutations exhibit a reduced tendency to aggregate during immobilization on an affinity matrix. Several of the suppressors also exhibit allele-specific behavior indicative of specific intramolecular interactions in the receptor.
Figures






Similar articles
-
Comparison of Experimental Approaches Used to Determine the Structure and Function of the Class D G Protein-Coupled Yeast α-Factor Receptor.Biomolecules. 2022 May 30;12(6):761. doi: 10.3390/biom12060761. Biomolecules. 2022. PMID: 35740886 Free PMC article. Review.
-
Identification of residues involved in homodimer formation located within a β-strand region of the N-terminus of a Yeast G protein-coupled receptor.J Recept Signal Transduct Res. 2012 Apr;32(2):65-75. doi: 10.3109/10799893.2011.647352. Epub 2012 Jan 24. J Recept Signal Transduct Res. 2012. PMID: 22268895
-
Identification of residues that contribute to receptor activation through the analysis of compensatory mutations in the G protein-coupled alpha-factor receptor.Biochemistry. 2005 Feb 1;44(4):1278-87. doi: 10.1021/bi048050u. Biochemistry. 2005. PMID: 15667221
-
Quantification of mutation-derived bias for alternate mating functionalities of the Saccharomyces cerevisiae Ste2p pheromone receptor.J Biochem. 2016 Jan;159(1):49-58. doi: 10.1093/jb/mvv072. Epub 2015 Jul 30. J Biochem. 2016. PMID: 26232403 Free PMC article.
-
Intensive mutational analysis of G protein-coupled receptors in yeast.Methods Mol Biol. 2004;237:105-20. doi: 10.1385/1-59259-430-1:105. Methods Mol Biol. 2004. PMID: 14501043 Review.
Cited by
-
Functional plasticity and evolutionary adaptation of allosteric regulation.Proc Natl Acad Sci U S A. 2020 Oct 13;117(41):25445-25454. doi: 10.1073/pnas.2002613117. Epub 2020 Sep 30. Proc Natl Acad Sci U S A. 2020. PMID: 32999067 Free PMC article.
-
Comparison of Experimental Approaches Used to Determine the Structure and Function of the Class D G Protein-Coupled Yeast α-Factor Receptor.Biomolecules. 2022 May 30;12(6):761. doi: 10.3390/biom12060761. Biomolecules. 2022. PMID: 35740886 Free PMC article. Review.
-
Analysis of random PCR-originated mutants of the yeast Ste2 and Ste3 receptors.Microbiologyopen. 2016 Aug;5(4):670-86. doi: 10.1002/mbo3.361. Epub 2016 May 5. Microbiologyopen. 2016. PMID: 27150158 Free PMC article.
-
A Paradigm for Peptide Hormone-GPCR Analyses.Molecules. 2020 Sep 18;25(18):4272. doi: 10.3390/molecules25184272. Molecules. 2020. PMID: 32961885 Free PMC article. Review.
-
Heterotrimeric G Protein-coupled Receptor Signaling in Yeast Mating Pheromone Response.J Biol Chem. 2016 Apr 8;291(15):7788-95. doi: 10.1074/jbc.R116.714980. Epub 2016 Feb 23. J Biol Chem. 2016. PMID: 26907689 Free PMC article. Review.
References
-
- Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nature Reviews Drug Discovery. 2006;5:993–996. - PubMed
-
- White SH. Biophysical dissection of membrane proteins. Nature. 2009;459:344–346. - PubMed
-
- Lundstrom K. Structural biology of G protein-coupled receptors. Bioorganic & Medicinal Chemistry Letters. 2005;15:3654–3657. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

