A sorghum MYB transcription factor induces 3-deoxyanthocyanidins and enhances resistance against leaf blights in maize
- PMID: 25647576
- PMCID: PMC6272393
- DOI: 10.3390/molecules20022388
A sorghum MYB transcription factor induces 3-deoxyanthocyanidins and enhances resistance against leaf blights in maize
Abstract
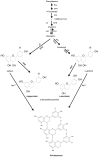
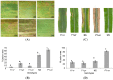
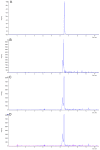
Sorghum responds to the ingress of the fungal pathogen Colletotrichum sublineolum through the biosynthesis of 3-deoxyanthocyanidin phytoalexins at the site of primary infection. Biosynthesis of 3-deoxyanthocyanidins in sorghum requires a MYB transcription factor encoded by yellow seed1 (y1), an orthologue of the maize gene pericarp color1 (p1). Maize lines with a functional p1 and flavonoid structural genes do not produce foliar 3-deoxyanthocyanidins in response to fungal ingress. To perform a comparative metabolic analysis of sorghum and maize 3-deoxyanthocyanidin biosynthetic pathways, we developed transgenic maize lines expressing the sorghum y1 gene. In maize, the y1 transgene phenocopied p1-regulated pigment accumulation in the pericarp and cob glumes. LC-MS profiling of fungus-challenged Y1-maize leaves showed induction of 3-deoxyanthocyanidins, specifically luteolinidin. Y1-maize plants also induced constitutive and higher levels of flavonoids in leaves. In response to Colletotrichum graminicola, Y1-maize showed a resistance response.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

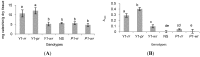
References
-
- Kristensen C., Morant M., Olsen C.E., Ekstrøm C.T., Galbraith D.W., Møller B.L., Bak S. Metabolic engineering of dhurrin in transgenic arabidopsis plants with marginal inadvertent effects on the metabolome and transcriptome. Proc. Natl. Acad. Sci. USA. 2005;102:1779–1784. doi: 10.1073/pnas.0409233102. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

