Isofunctional enzymes PAD1 and UbiX catalyze formation of a novel cofactor required by ferulic acid decarboxylase and 4-hydroxy-3-polyprenylbenzoic acid decarboxylase
- PMID: 25647642
- PMCID: PMC11756644
- DOI: 10.1021/cb5008103
Isofunctional enzymes PAD1 and UbiX catalyze formation of a novel cofactor required by ferulic acid decarboxylase and 4-hydroxy-3-polyprenylbenzoic acid decarboxylase
Abstract
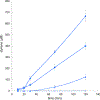
The decarboxylation of antimicrobial aromatic acids such as phenylacrylic acid (cinnamic acid) and ferulic acid by yeast requires two enzymes described as phenylacrylic acid decarboxylase (PAD1) and ferulic acid decarboxylase (FDC). These enzymes are of interest for various biotechnological applications, such as the production of chemical feedstocks from lignin under mild conditions. However, the specific role of each protein in catalyzing the decarboxylation reaction remains unknown. To examine this, we have overexpressed and purified both PAD1 and FDC from E. coli. We demonstrate that PAD1 is a flavin mononucleotide (FMN)-containing protein. However, it does not function as a decarboxylase. Rather, PAD1 catalyzes the formation of a novel, diffusible cofactor required by FDC for decarboxylase activity. Coexpression of FDC and PAD1 results in the production of FDC with high levels cofactor bound. Holo-FDC catalyzes the decarboxylation of phenylacrylic acid, coumaric acid and ferulic acid with apparent kcat ranging from 1.4-4.6 s(-1). The UV-visible and mass spectra of the cofactor indicate that it appears to be a novel, modified form of reduced FMN; however, its instability precluded determination of its structure. The E. coli enzymes UbiX and UbiD are related by sequence to PAD1 and FDC respectively and are involved in the decarboxylation of 4-hydroxy-3-octaprenylbenzoic acid, an intermediate in ubiquinone biosynthesis. We found that endogenous UbiX can also activate FDC. This implies that the same cofactor is required for decarboxylation of 4-hydroxy-3-polyprenylbenzoic acid by UbiD and suggests a wider role for this cofactor in metabolism.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
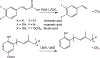






References
-
- Cleland WW (1999) Mechanisms of enzymatic oxidative decarboxylation. Acc. Chem. Res. 32, 862–868.
-
- Belcher J, McLean KJ, Matthews S, Woodward LS, Fisher K, Rigby SEJ, Nelson DR, Potts D, Baynham MT, Parker DA, Leys D, and Munro AW (2014) Structure and biochemical properties of the alkene producing cytochrome P450 OleTJE (CYP152L1) from the Jeotgalicoccus sp. 8456 bacterium. J. Biol. Chem. 289, 6535–6550. - PMC - PubMed
-
- Kourist R, Guterl J-K, Miyamoto K, and Sieber V (2014) Enzymatic decarboxylation—An emerging reaction for chemicals production from renewable resources. ChemCatChem 6, 689–701.
-
- Straathof AJJ (2014) Transformation of biomass into commodity chemicals using enzymes or cells. Chem. Rev. 114, 1871–1908. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

