Identification of fragments from Autographa californica polyhedrin protein essential for self-aggregation and exogenous protein incorporation
- PMID: 25648249
- PMCID: PMC4320575
- DOI: 10.1186/s12858-015-0034-9
Identification of fragments from Autographa californica polyhedrin protein essential for self-aggregation and exogenous protein incorporation
Abstract
Background: Baculoviruses are widely used for the production of recombinant proteins, biopesticides and as gene delivery systems. One of the viral forms called polyhedra has been recently exploited as a scaffold system to incorporate or encapsulate foreign proteins or peptide fragments. However, an efficient strategy for foreign protein incorporation has not been thoroughly studied.
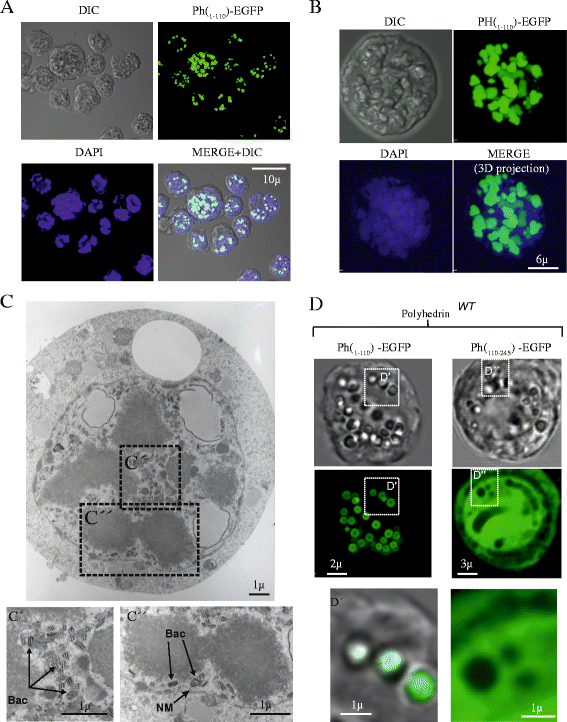
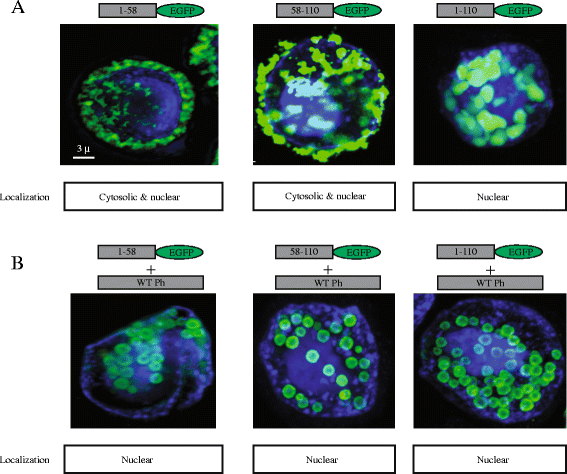
Results: Based on the crystal structure of polyhedrin, we conducted an in silico analysis of the baculovirus Autographa californica nucleopolyhedrovirus (AcMNPV) polyhedrin protein to select the minimum fragments of polyhedrin that could be incorporated into polyhedra. Using confocal and transmission electron microscopy we analyzed the expression and cellular localization of the different polyhedrin fragments fused to the green fluorescent protein (EGFP) used as reporter. The amino fragment 1-110 contains two repeats formed each of two β sheets followed by a α helix (amino acids 1-58 and 58-110) that are important for the formation and stability of polyhedra. These fragments 1-58, 58-110 and 1-110 could be incorporated into polyhedra. However, only fragments 1-110 and 58-110 can self-aggregate.
Conclusions: These results demonstrate that 58-110 is the minimum fragment that contributes to the assembly of the recombinant polyhedra via self-aggregation. This is the minimum sequence that can be used to efficiently incorporate foreign proteins into polyhedra.
Figures





Similar articles
-
Immobilization of foreign protein in BmNPV polyhedra by fusion expression with partial polyhedrin fragments.J Virol Methods. 2013 Dec;194(1-2):185-9. doi: 10.1016/j.jviromet.2013.08.020. Epub 2013 Sep 2. J Virol Methods. 2013. PMID: 24008009
-
Improving Baculovirus Infectivity by Efficiently Embedding Enhancing Factors into Occlusion Bodies.Appl Environ Microbiol. 2017 Jun 30;83(14):e00595-17. doi: 10.1128/AEM.00595-17. Print 2017 Jul 15. Appl Environ Microbiol. 2017. PMID: 28500037 Free PMC article.
-
Characterization of a new Autographa californica multiple nucleopolyhedrovirus (AcMNPV) polyhedra mutant.Virus Res. 2009 Mar;140(1-2):1-7. doi: 10.1016/j.virusres.2008.10.010. Epub 2008 Dec 18. Virus Res. 2009. PMID: 19038296
-
Comparison between different conditions for the incorporation of foreign proteins into Autographa californica multiple polyhedrovirus polyhedra for biotechnological purposes.Arch Virol. 2024 Apr 24;169(5):108. doi: 10.1007/s00705-024-06015-5. Arch Virol. 2024. PMID: 38658418
-
Biotechnological applications of occlusion bodies of Baculoviruses.Appl Microbiol Biotechnol. 2018 Aug;102(16):6765-6774. doi: 10.1007/s00253-018-9130-2. Epub 2018 Jun 5. Appl Microbiol Biotechnol. 2018. PMID: 29872886 Review.
Cited by
-
A self-aggregating peptide: implications for the development of thermostable vaccine candidates.BMC Biotechnol. 2020 Jan 21;20(1):1. doi: 10.1186/s12896-019-0592-9. BMC Biotechnol. 2020. PMID: 31959159 Free PMC article.
-
Exploiting Nanomedicine for Cancer Polychemotherapy: Recent Advances and Clinical Applications.Pharmaceutics. 2023 Mar 14;15(3):937. doi: 10.3390/pharmaceutics15030937. Pharmaceutics. 2023. PMID: 36986798 Free PMC article. Review.
-
Recombinant occlusion bodies of baculovirus as carriers of a non-structural protein of foot-and-mouth disease virus.3 Biotech. 2018 Nov;8(11):457. doi: 10.1007/s13205-018-1482-x. Epub 2018 Oct 20. 3 Biotech. 2018. PMID: 30370198 Free PMC article.
-
An ambient-temperature stable nanoparticle-based vaccine for nasal application that confers long-lasting immunogenicity to carried antigens.Front Immunol. 2022 Oct 31;13:1057499. doi: 10.3389/fimmu.2022.1057499. eCollection 2022. Front Immunol. 2022. PMID: 36389760 Free PMC article.
-
Easily purified baculovirus/insect-system-expressed recombinant hepatitis B virus surface antigen fused to the N- or C-terminus of polyhedrin.Arch Virol. 2022 Feb;167(2):345-354. doi: 10.1007/s00705-021-05305-6. Epub 2021 Nov 28. Arch Virol. 2022. PMID: 34839419
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

