xiNET: cross-link network maps with residue resolution
- PMID: 25648531
- PMCID: PMC4390258
- DOI: 10.1074/mcp.O114.042259
xiNET: cross-link network maps with residue resolution
Abstract
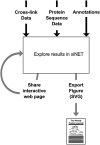
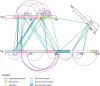
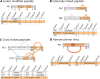
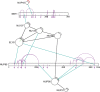
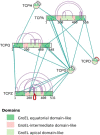
xiNET is a visualization tool for exploring cross-linking/mass spectrometry results. The interactive maps of the cross-link network that it generates are a type of node-link diagram. In these maps xiNET displays: (1) residue resolution positional information including linkage sites and linked peptides; (2) all types of cross-linking reaction product; (3) ambiguous results; and, (4) additional sequence information such as domains. xiNET runs in a browser and exports vector graphics which can be edited in common drawing packages to create publication quality figures.
Availability: xiNET is open source, released under the Apache version 2 license. Results can be viewed by uploading data to http://crosslinkviewer.org/ or by downloading the software from http://github.com/colin-combe/crosslink-viewer and running it locally.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



References
-
- Herzog F., Kahraman A., Boehringer D., Mak R., Bracher A., Walzthoeni T., Leitner A., Beck M., Hartl F. U., Ban N., Malmström L., Aebersold R. (2012) Structural probing of a protein phosphatase 2a network by chemical cross-linking and mass spectrometry. Science 337, 1348–1352 - PubMed
-
- Gehlenborg N., Wong B. (2012) Points of view: Networks. Nat. Methods 9, 115. - PubMed
-
- Seebacher J., Mallick P., Zhang N., Eddes J. S., Aebersold R., Gelb M. H. (2006) Protein cross-linking analysis using mass spectrometry, isotope-coded cross-linkers, and integrated computational data processing. J. Proteome Res. 5, 2270–2282 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

