The intracellular domain of teneurin-1 induces the activity of microphthalmia-associated transcription factor (MITF) by binding to transcriptional repressor HINT1
- PMID: 25648896
- PMCID: PMC4375472
- DOI: 10.1074/jbc.M114.615922
The intracellular domain of teneurin-1 induces the activity of microphthalmia-associated transcription factor (MITF) by binding to transcriptional repressor HINT1
Abstract
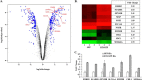
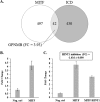
Teneurins are large type II transmembrane proteins that are necessary for the normal development of the CNS. Although many studies highlight the significance of teneurins, especially during development, there is only limited information known about the molecular mechanisms of function. Previous studies have shown that the N-terminal intracellular domain (ICD) of teneurins can be cleaved at the membrane and subsequently translocates to the nucleus, where it can influence gene transcription. Because teneurin ICDs do not contain any intrinsic DNA binding sequences, interaction partners are required to affect transcription. Here, we identified histidine triad nucleotide binding protein 1 (HINT1) as a human teneurin-1 ICD interaction partner in a yeast two-hybrid screen. This interaction was confirmed in human cells, where HINT1 is known to inhibit the transcription of target genes by directly binding to transcription factors at the promoter. In a whole transcriptome analysis of BS149 glioblastoma cells overexpressing the teneurin-1 ICD, several microphthalmia-associated transcription factor (MITF) target genes were found to be up-regulated. Directly comparing the transcriptomes of MITF versus TEN1-ICD-overexpressing BS149 cells revealed 42 co-regulated genes, including glycoprotein non-metastatic b (GPNMB). Using real-time quantitative PCR to detect endogenous GPNMB expression upon overexpression of MITF and HINT1 as well as promoter reporter assays using GPNMB promoter constructs, we could demonstrate that the teneurin-1 ICD binds HINT1, thus switching on MITF-dependent transcription of GPNMB.
Keywords: BEX1; GPNMB; Glioblastoma; MACF1; Microarray; ODZ; Protein-Protein Interaction; Receptor; TENM1; Transcription Repressor.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



References
-
- Baumgartner S., Chiquet-Ehrismann R. (1993) Tena, a Drosophila gene related to tenascin, shows selective transcript localization. Mech. Dev. 40, 165–176 - PubMed
-
- Levine A., Bashan-Ahrend A., Budai-Hadrian O., Gartenberg D., Menasherow S., Wides R. (1994) Odd Oz: a novel Drosophila pair rule gene. Cell 77, 587–598 - PubMed
-
- Minet A. D., Chiquet-Ehrismann R. (2000) Phylogenetic analysis of teneurin genes and comparison to the rearrangement hot spot elements of E. coli. Gene 257, 87–97 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

