Global analysis of the role of autophagy in cellular metabolism and energy homeostasis in Arabidopsis seedlings under carbon starvation
- PMID: 25649436
- PMCID: PMC4456922
- DOI: 10.1105/tpc.114.134205
Global analysis of the role of autophagy in cellular metabolism and energy homeostasis in Arabidopsis seedlings under carbon starvation
Abstract
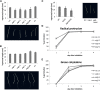
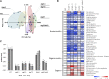
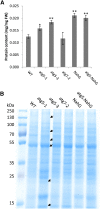
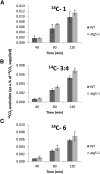
Germination and early seedling establishment are developmental stages in which plants face limited nutrient supply as their photosynthesis mechanism is not yet active. For this reason, the plant must mobilize the nutrient reserves provided by the mother plant in order to facilitate growth. Autophagy is a catabolic process enabling the bulk degradation of cellular constituents in the vacuole. The autophagy mechanism is conserved among eukaryotes, and homologs of many autophagy-related (ATG) genes have been found in Arabidopsis thaliana. T-DNA insertion mutants (atg mutants) of these genes display higher sensitivity to various stresses, particularly nutrient starvation. However, the direct impact of autophagy on cellular metabolism has not been well studied. In this work, we used etiolated Arabidopsis seedlings as a model system for carbon starvation. atg mutant seedlings display delayed growth in response to carbon starvation compared with wild-type seedlings. High-throughput metabolomic, lipidomic, and proteomic analyses were performed, as well as extensive flux analyses, in order to decipher the underlying causes of the phenotype. Significant differences between atg mutants and wild-type plants have been demonstrated, suggesting global effects of autophagy on central metabolism during carbon starvation as well as severe energy deprivation, resulting in a morphological phenotype.
© 2015 American Society of Plant Biologists. All rights reserved.
Figures


References
-
- Araújo W.L., Ishizaki K., Nunes-Nesi A., Larson T.R., Tohge T., Krahnert I., Witt S., Obata T., Schauer N., Graham I.A., Leaver C.J., Fernie A.R. (2010). Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell 22: 1549–1563. - PMC - PubMed
-
- Araújo W.L., Tohge T., Ishizaki K., Leaver C.J., Fernie A.R. (2011). Protein degradation - an alternative respiratory substrate for stressed plants. Trends Plant Sci. 16: 489–498. - PubMed
-
- Arc E., Galland M., Cueff G., Godin B., Lounifi I., Job D., Rajjou L. (2011). Reboot the system thanks to protein post-translational modifications and proteome diversity: How quiescent seeds restart their metabolism to prepare seedling establishment. Proteomics 11: 1606–1618. - PubMed
-
- Avin-Wittenberg T., Fernie A.R. (2014). At long last: evidence for pexophagy in plants. Mol. Plant 7: 1257–1260. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

