The membrane proximal external regions of gp41 from HIV-1 strains HXB2 and JRFL have different sensitivities to alanine mutation
- PMID: 25649507
- PMCID: PMC4684184
- DOI: 10.1021/bi501171r
The membrane proximal external regions of gp41 from HIV-1 strains HXB2 and JRFL have different sensitivities to alanine mutation
Abstract
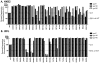
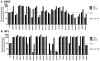
The transmembrane subunit (gp41) of the HIV envelope protein complex (Env) mediates the viral fusion step of HIV entry. The membrane proximal external region (MPER), one of the functional domains of gp41, has been the focus of a great deal of research because it is a target for neutralizing antibodies. In this study, we examined 23 amino acid residues in the MPER (660-683) in both a CXCR4 coreceptor-utilizing strain (HXB2) and a CCR5-utilizing strain (JRFL) by alanine scanning mutagenesis. Despite the high degree of gp41 sequence conservation, the effects of alanine mutation in the MPER were different between the two strains. Most mutations in HXB2 had fusogenicity and protein expression levels not less than 50% of that of the wild type in the case of cell-cell fusion. However, ∼30% of the mutants in HXB2 showed a severe defect in fusogenicity in viral entry. Mutations in the MPER of strain JRFL had more dramatic effects than that in HXB2 in cell-cell fusion and viral entry. The fact that there are large differences in the effects of mutation between two strains suggests the potential for the interaction of the MPER with nonconserved sequences such as the fusion peptide and/or other NHR domains as well as potential long-range structural effects on the conformational changes that occur with the Env complex during membrane fusion.
Figures

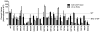

References
-
- Montero M, van Houten NE, Wang X, Scott JK. The membrane-proximal external region of the human immunodeficiency virus type 1 envelope: dominant site of antibody neutralization and target for vaccine design. Microbiology and molecular biology reviews : MMBR. 2008;72:54–84. table of contents. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

