Reference-based compression of short-read sequences using path encoding
- PMID: 25649622
- PMCID: PMC4481695
- DOI: 10.1093/bioinformatics/btv071
Reference-based compression of short-read sequences using path encoding
Abstract
Motivation: Storing, transmitting and archiving data produced by next-generation sequencing is a significant computational burden. New compression techniques tailored to short-read sequence data are needed.
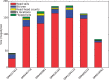
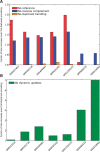
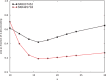
Results: We present here an approach to compression that reduces the difficulty of managing large-scale sequencing data. Our novel approach sits between pure reference-based compression and reference-free compression and combines much of the benefit of reference-based approaches with the flexibility of de novo encoding. Our method, called path encoding, draws a connection between storing paths in de Bruijn graphs and context-dependent arithmetic coding. Supporting this method is a system to compactly store sets of kmers that is of independent interest. We are able to encode RNA-seq reads using 3-11% of the space of the sequence in raw FASTA files, which is on average more than 34% smaller than competing approaches. We also show that even if the reference is very poorly matched to the reads that are being encoded, good compression can still be achieved.
Availability and implementation: Source code and binaries freely available for download at http://www.cs.cmu.edu/∼ckingsf/software/pathenc/, implemented in Go and supported on Linux and Mac OS X.
© The Author 2015. Published by Oxford University Press.
Figures
References
-
- Adjeroh D., et al. (2002) DNA sequence compression using the Burrows-Wheeler transform. In: Procceeding IEEE Computer Society Bioinformatics Conference. Vol. 1, IEEE Computer Society, Washington, DC, pp. 303–313. - PubMed
-
- Bhola V., et al. (2011) No-reference compression of genomic data stored in FASTQ format. In: IEEE International Conference on Bioinformatics and Biomedicine. IEEE Computer Society, Washington, DC, pp. 147–150
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

