Expanding the enzyme universe: accessing non-natural reactions by mechanism-guided directed evolution
- PMID: 25649694
- PMCID: PMC4404643
- DOI: 10.1002/anie.201409470
Expanding the enzyme universe: accessing non-natural reactions by mechanism-guided directed evolution
Abstract
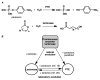
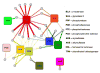
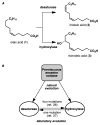
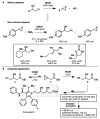
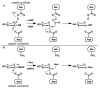
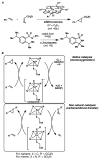
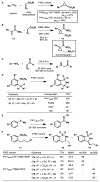
High selectivity and exquisite control over the outcome of reactions entice chemists to use biocatalysts in organic synthesis. However, many useful reactions are not accessible because they are not in nature's known repertoire. In this Review, we outline an evolutionary approach to engineering enzymes to catalyze reactions not found in nature. We begin with examples of how nature has discovered new catalytic functions and how such evolutionary progression has been recapitulated in the laboratory starting from extant enzymes. We then examine non-native enzyme activities that have been exploited for chemical synthesis, with an emphasis on reactions that do not have natural counterparts. Non-natural activities can be improved by directed evolution, thus mimicking the process used by nature to create new catalysts. Finally, we describe the discovery of non-native catalytic functions that may provide future opportunities for the expansion of the enzyme universe.
Keywords: biocatalysis; enzymes; non-natural activity; promiscuity; protein engineering.
© 2015 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures



References
-
- Moore JC, Arnold FH. Nat Biotechnol. 1996;14:458–467. - PubMed
- Shao Z, Arnold FH. Curr Opin Struct Biol. 1996;6:513–518. - PubMed
- Arnold FH. Chem Eng Sci. 1996;51:5091–5102.
- Arnold FH, Moore JC. Adv Biochem Eng Biotechnol. 1997;58:1–14. - PubMed
- Reetz MT, Zonta A, Schimossek K, Liebeton K, Jaeger K-E. Angew Chem Int Ed. 1997;36:2830–2832.
- Reetz MT. J Am Chem Soc. 2013;135:12480–12496. - PubMed
-
- Savile CK, Janey JM, Mundorff EC, Moore JC, Tam S, Jarvis WR, Colbeck JC, Krebber A, Fleitz FJ, Brands J, Devine PN, Huisman GW, Hughes GJ. Science. 2010;329:305–308. - PubMed
- Liang J, Lalonde J, Borup B, Mitchell V, Mundorff E, Trinh N, Kochrekar DA, Cherat RN, Ganesh Pai G. Org Proc Res Dev. 2010;14:193–198.
- Bornscheuer UT, Huisman GW, Kazlauskas RJ, Lutz S, Moore JC, Robins K. Nature. 2012;485:185–194. - PubMed
- Alexeeva M, Enright A, Dawson MJ, Mahmoudian M, Turner NJ. Angew Chem Int Ed. 2002;41:3177–3180. - PubMed
- Turner NJ. Nat Chem Biol. 2009;5:567–573. - PubMed
-
- Wohlgemuth R. Curr Opin Biotechnol. 2010;21:713–724. - PubMed
-
- Steen EJ, Kang Y, Bokinsky G, Hu Z, Schirmer A, McClure A, del Cardayre SB, Keasling JD. Nature. 2010;463:559–563. - PubMed
- Peralta-Yahya PP, Zhang F, del Cardayre SB, Keasling JD. Nature. 2012;488:320–328. - PubMed
- Atsumi S, Hanai T, Liao JC. Nature. 2008;451:86–89. - PubMed
- Lee SY, Park JH, Jang SH, Nielsen LK, Kim J, Jung KS. Biotechnol Bioeng. 2008;101:209–228. - PubMed
-
- Jansen RA. Annu Rev Microbiol. 1976;30:409–425. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

