Identifying genetic interactions associated with late-onset Alzheimer's disease
- PMID: 25649863
- PMCID: PMC4300162
- DOI: 10.1186/s13040-014-0035-z
Identifying genetic interactions associated with late-onset Alzheimer's disease
Abstract
Background: Identifying genetic interactions in data obtained from genome-wide association studies (GWASs) can help in understanding the genetic basis of complex diseases. The large number of single nucleotide polymorphisms (SNPs) in GWASs however makes the identification of genetic interactions computationally challenging. We developed the Bayesian Combinatorial Method (BCM) that can identify pairs of SNPs that in combination have high statistical association with disease.
Results: We applied BCM to two late-onset Alzheimer's disease (LOAD) GWAS datasets to identify SNPs that interact with known Alzheimer associated SNPs. We also compared BCM with logistic regression that is implemented in PLINK. Gene Ontology analysis of genes from the top 200 dataset SNPs for both GWAS datasets showed overrepresentation of LOAD-related terms. Four genes were common to both datasets: APOE and APOC1, which have well established associations with LOAD, and CAMK1D and FBXL13, not previously linked to LOAD but having evidence of involvement in LOAD. Supporting evidence was also found for additional genes from the top 30 dataset SNPs.
Conclusion: BCM performed well in identifying several SNPs having evidence of involvement in the pathogenesis of LOAD that would not have been identified by univariate analysis due to small main effect. These results provide support for applying BCM to identify potential genetic variants such as SNPs from high dimensional GWAS datasets.
Keywords: Alzheimer’s disease; Bayesian networks; Epistasis; Genome-wide association study.
Figures
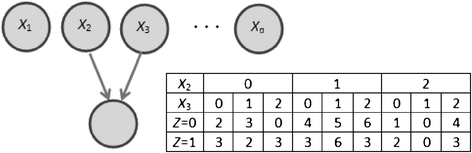
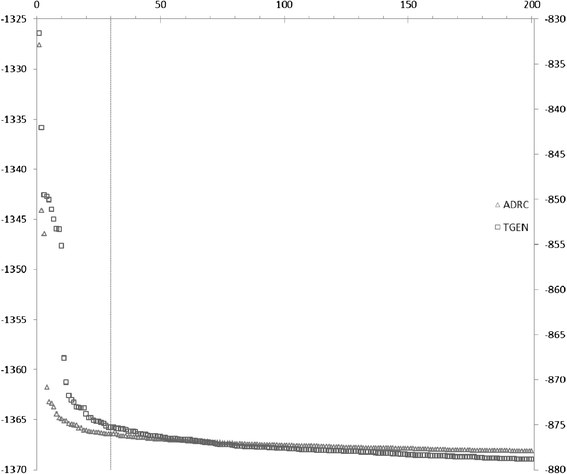
Similar articles
-
LEAP: biomarker inference through learning and evaluating association patterns.Genet Epidemiol. 2015 Mar;39(3):173-84. doi: 10.1002/gepi.21889. Epub 2015 Feb 12. Genet Epidemiol. 2015. PMID: 25677188 Free PMC article.
-
The application of network label propagation to rank biomarkers in genome-wide Alzheimer's data.BMC Genomics. 2014 Apr 14;15:282. doi: 10.1186/1471-2164-15-282. BMC Genomics. 2014. PMID: 24731236 Free PMC article.
-
Identification of a late onset Alzheimer's disease candidate risk variant at 9q21.33 in Polish patients.J Alzheimers Dis. 2012;32(1):157-68. doi: 10.3233/JAD-2012-120520. J Alzheimers Dis. 2012. PMID: 22785395
-
Shared genetic etiology underlying Alzheimer's disease and type 2 diabetes.Mol Aspects Med. 2015 Jun-Oct;43-44:66-76. doi: 10.1016/j.mam.2015.06.006. Epub 2015 Jun 23. Mol Aspects Med. 2015. PMID: 26116273 Free PMC article. Review.
-
[Personal genomics for Alzheimer's disease].Brain Nerve. 2013 Mar;65(3):235-46. Brain Nerve. 2013. PMID: 23475515 Review. Japanese.
Cited by
-
The interaction of early life factors and depression-associated loci affecting the age at onset of the depression.Transl Psychiatry. 2022 Jul 25;12(1):294. doi: 10.1038/s41398-022-02042-5. Transl Psychiatry. 2022. PMID: 35879288 Free PMC article.
-
Alzheimer's disease-associated U1 snRNP splicing dysfunction causes neuronal hyperexcitability and cognitive impairment.Nat Aging. 2022 Oct;2(10):923-940. doi: 10.1038/s43587-022-00290-0. Epub 2022 Oct 12. Nat Aging. 2022. PMID: 36636325 Free PMC article.
-
Navigating the blurred boundary: Neuropathologic changes versus clinical symptoms in Alzheimer's disease, and its consequences for research in genetics.J Alzheimers Dis. 2025 Apr;104(3):611-626. doi: 10.1177/13872877251317543. Epub 2025 Feb 16. J Alzheimers Dis. 2025. PMID: 39956949 Free PMC article. Review.
-
LEAP: biomarker inference through learning and evaluating association patterns.Genet Epidemiol. 2015 Mar;39(3):173-84. doi: 10.1002/gepi.21889. Epub 2015 Feb 12. Genet Epidemiol. 2015. PMID: 25677188 Free PMC article.
-
GSAP regulates lipid homeostasis and mitochondrial function associated with Alzheimer's disease.J Exp Med. 2021 Aug 2;218(8):e20202446. doi: 10.1084/jem.20202446. Epub 2021 Jun 22. J Exp Med. 2021. PMID: 34156424 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

