Evidence for a sexual dimorphism in gene expression noise in metazoan species
- PMID: 25649874
- PMCID: PMC4314086
- DOI: 10.7717/peerj.750
Evidence for a sexual dimorphism in gene expression noise in metazoan species
Abstract
Many biological processes depend on very few copies of intervening elements, which makes such processes particularly susceptible to the stochastic fluctuations of these elements. The intrinsic stochasticity of certain processes is propagated across biological levels, causing genotype- and environment-independent biological variation which might permit populations to better cope with variable environments. Biological variations of stochastic nature might also allow the accumulation of variations at the genetic level that are hidden from natural selection, which might have a great potential for population diversification. The study of any mechanism that resulted in the modulation of stochastic variation is, therefore, of potentially wide interest. I propose that sex might be an important modulator of the stochastic variation in gene expression, i.e., gene expression noise. Based on known associations between different patterns of gene expression variation, I hypothesize that in metazoans the gene expression noise might be generally larger in heterogametic than in homogametic individuals. I directly tested this hypothesis by comparing putative genotype- and environment-independent variations in gene expression between females and males of Drosophila melanogaster strains. Also, considering the potential effect of the propagation of gene expression noise across biological levels, I indirectly tested the existence of a metazoan sexual dimorphism in gene expression noise by analyzing putative genotype- and environment-independent variation in phenotypes related to interaction with the environment in D. melanogaster strains and metazoan species. The results of these analyses are consistent with the hypothesis that gene expression is generally noisier in heterogametic than in homogametic individuals. Further analyses and discussion of existing literature permits the speculation that the sexual dimorphism in gene expression noise is ultimately based on the nuclear dynamics in gametogenesis and very early embryogenesis of sex-specific chromosomes, i.e., Y and W chromosomes.
Keywords: Gene expression noise; Genomic tuning knobs; Heterochromatin; Sex; Sex-specific chromosomes.
Conflict of interest statement
The author declares there are no competing interests.
Figures

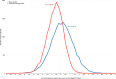
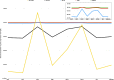

Similar articles
-
Effect of genomic deficiencies on sexual size dimorphism through modification of developmental time in Drosophila melanogaster.Heredity (Edinb). 2015 Aug;115(2):140-5. doi: 10.1038/hdy.2015.1. Epub 2015 Apr 22. Heredity (Edinb). 2015. PMID: 25899012 Free PMC article.
-
Gene duplication in the evolution of sexual dimorphism.Evolution. 2012 May;66(5):1556-66. doi: 10.1111/j.1558-5646.2011.01525.x. Epub 2012 Jan 12. Evolution. 2012. PMID: 22519790
-
Quantitative genetics of natural variation of behavior in Drosophila melanogaster: the possible role of the social environment on creating persistent patterns of group activity.Evolution. 2005 Jul;59(7):1529-39. Evolution. 2005. PMID: 16153038
-
Sexual dimorphism in flowering plants.J Exp Bot. 2013 Jan;64(1):67-82. doi: 10.1093/jxb/ers308. Epub 2012 Nov 25. J Exp Bot. 2013. PMID: 23183260 Review.
-
Modeling cell-to-cell stochastic variability in intrinsic apoptosis pathway.Annu Int Conf IEEE Eng Med Biol Soc. 2012;2012:5498-501. doi: 10.1109/EMBC.2012.6347239. Annu Int Conf IEEE Eng Med Biol Soc. 2012. PMID: 23367174 Review.
Cited by
-
Transgenerational Self-Reconstruction of Disrupted Chromatin Organization After Exposure To An Environmental Stressor in Mice.Sci Rep. 2019 Sep 10;9(1):13057. doi: 10.1038/s41598-019-49440-2. Sci Rep. 2019. PMID: 31506492 Free PMC article.
-
Transcriptome dynamics along axolotl regenerative development are consistent with an extensive reduction in gene expression heterogeneity in dedifferentiated cells.PeerJ. 2017 Nov 6;5:e4004. doi: 10.7717/peerj.4004. eCollection 2017. PeerJ. 2017. PMID: 29134148 Free PMC article.
-
Ancestral perinatal obesogen exposure results in a transgenerational thrifty phenotype in mice.Nat Commun. 2017 Dec 8;8(1):2012. doi: 10.1038/s41467-017-01944-z. Nat Commun. 2017. PMID: 29222412 Free PMC article.
-
Transcriptional Interference Promotes Rapid Expression Divergence of Drosophila Nested Genes.Genome Biol Evol. 2016 Oct 23;8(10):3149-3158. doi: 10.1093/gbe/evw237. Genome Biol Evol. 2016. PMID: 27664180 Free PMC article.
References
-
- Ardila-Garcia AM, Gregory TR. An exploration of genome size diversity in dragonflies and damselflies (Insecta: Odonata) Journal of Zoology. 2009;278:163–173. doi: 10.1111/j.1469-7998.2009.00557.x. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

