Mutated Ptpn11 alters leukemic stem cell frequency and reduces the sensitivity of acute myeloid leukemia cells to Mcl1 inhibition
- PMID: 25650089
- PMCID: PMC4456293
- DOI: 10.1038/leu.2015.18
Mutated Ptpn11 alters leukemic stem cell frequency and reduces the sensitivity of acute myeloid leukemia cells to Mcl1 inhibition
Abstract
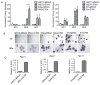
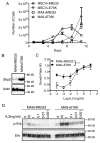
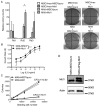
PTPN11 encodes the Shp2 non-receptor protein-tyrosine phosphatase implicated in several signaling pathways. Activating mutations in Shp2 are commonly associated with juvenile myelomonocytic leukemia but are not as well defined in other neoplasms. Here we report that Shp2 mutations occur in human acute myeloid leukemia (AML) at a rate of 6.6% (6/91) in the ECOG E1900 data set. We examined the role of mutated Shp2 in leukemias harboring MLL translocations, which co-occur in human AML. The hyperactive Shp2E76K mutant, commonly observed in leukemia patients, significantly accelerated MLL-AF9-mediated leukemogenesis in vivo. Shp2E76K increased leukemic stem cell frequency and affords MLL-AF9 leukemic cells IL3 cytokine hypersensitivity. As Shp2 is reported to regulate anti-apoptotic genes, we investigated Bcl2, Bcl-xL and Mcl1 expression in MLL-AF9 leukemic cells with and without Shp2E76K. Although the Bcl2 family of genes was upregulated in Shp2E76K cells, Mcl1 showed the highest upregulation in MLL-AF9 cells in response to Shp2E76K. Indeed, expression of Mcl1 in MLL-AF9 cells phenocopies expression of Shp2E76K, suggesting Shp2 mutations cooperate through activation of anti-apoptotic genes. Finally, we show Shp2E76K mutations reduce sensitivity of AML cells to small-molecule-mediated Mcl1 inhibition, suggesting reduced efficacy of drugs targeting MCL1 in patients with hyperactive Shp2.
Conflict of interest statement
The authors declare no conflict of interest.
The authors have no competing financial interests.
Figures

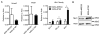

References
-
- Raimondi SC, Chang MN, Ravindranath Y, Behm FG, Gresik MV, Steuber CP, et al. Chromosomal abnormalities in 478 children with acute myeloid leukemia: clinical characteristics and treatment outcome in a cooperative pediatric oncology group study-POG 8821. Blood. 1999 Dec 1;94(11):3707–16. - PubMed
-
- Rubnitz JE, Link MP, Shuster JJ, Carroll AJ, Hakami N, Frankel LS, et al. Frequency and prognostic significance of HRX rearrangements in infant acute lymphoblastic leukemia: a Pediatric Oncology Group study. Blood. 1994 Jul 15;84(2):570–3. - PubMed
-
- Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet. 2007 Jan 1;16(1):92–106. Epub 2006/12/01. eng. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

