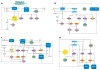
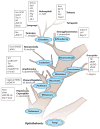
Figure 2
The basic TF network circuitry of T cell development and variations incorporated into it generate lymphoid effector cell types. Minimal components of each circuit are depicted. Cytokine receptor–signaling inputs controlling TF expression are not shown. (a) The foundation regulatory TF circuit of T cell development is the NOTCH-TCF1 hub (rectangles) that intimately integrates with the E2A regulatory domain and induces central genes (circles) necessary for T cell development from ETPs. This process operates in conjunction with TFs already expressed in LMPPs or CLPs that are active at multiple states of T cells (Ikaros, Myb, Runx1, Gfi1, Sox4, and others belong to this cluster). E2A induces the expression of IL-7R and RAG proteins required for the V(D)J recombination of Tcr genes. E2A and TCF1 control chromatin accessibility of the Tcr loci and regulate their ordered gene rearrangements. TCR signaling, along with E2A, can promote PLZF expression. But in adaptive T cells, PLZF is under an autoregulatory feedback loop involving ID3, induced by TCR signaling that in turn inhibits E2A to maintain a naive state of mature thymocytes. Absence of TCF1 in mice leads to a complete loss of T cell–committed DN2 precursor cells, and TCF1 controls the expression of one of the markers of DN2, IL-2R α-chain. TCR signaling, NOTCH, and TCF1 cooperate to turn on the effector lineage controlling TFs GATA3, LEF1, and RORγt (diamonds), but during intrathymic αβT cell development, they do not access the target cytokine gene loci. NOTCH and TCF1 target genes, most notably Bcl11b, inhibit genes active in non–T cell lymphoid lineages, such as ID2, which is required for ILE development. (b) Three sequentially incorporated ILE TFs, NFIL3 (nuclear factor, IL-3 regulated), ID2, and TOX (thymocyte selection–associated high-mobility group box), constitute the ILC hub. The high level of ID2 expression, induced by NFIL3, possibly in concert with TOX, leads to a muted activity of E2A that in turn impedes the cooperative NOTCH-E2A outputs. The consequences are lack of antigen receptor expression and varied dependence on NOTCH signaling for ILC differentiation. NFIL3 can inhibit Rorc expression in Th17 cells (dashed lines represent connections by function only; direct gene-to-gene interactions have not been confirmed in indicated cell types), and conversely, it turns on Eomes in NK cells to regulate IFN-γ (ovals, effector cytokines). In other ILC1s, T-bet, not EOMES, drives IFN-γ production. For ILC2s, TCF1 regulates Gata3 and cytokine receptor genes critical for ILC2 development. RORα is essential for ILC2 differentiation, but its upstream inducers and downstream targets are unmapped. PLZF is transiently induced, perhaps by RAS signaling in conjunction with E2A, and is also required for ILC2s, but its target genes are unmapped in ILCs. As in T cells, TCF1 inhibits IL-17 production and promotes IL-2R α-chain expression in ILC3s. TCF1 may contribute to Rorc transcription in ILCs directly, as in T cells, or indirectly, by promoting IL-7R expression, which in turn enhances RORγt expression (not depicted). RAR and other positive regulators of RORγt further entrench the ILC3 program. (c) Incorporation of two additional HMG TFs, SOX13 and SOX4, institutes the TCR+ ILE program. Activities of TCF1 are modulated by its interacting partners SOX13 and SOX4, which together turn on Rorc, with additional inputs from RAR and ETV5. SOX13 also induces Etv5, Blk, and Il7r and inhibits Lef1 to specify the Tγδ17 cell program. SOX4 regulates iNKT cell development, potentially by calibrating TCR signals, but the molecular mechanism remains to be ascertained. PLZF, ID3, and T-bet induction is initiated by TCR signaling. TCF1 positively regulates Gata3, to generate IL-4-producing cells, and Eomes and Lef1, to control IFN-γ synthesis. (d) A simplified adaptive Th effector circuit illustrates the dominance of TCR signaling as the arbiter of functional specialization. TFs belonging to the IRF-BATF axis, along with others induced by TCR and cytokine receptor signaling, constitute the hub. IRF4 and BATF cooperatively induce RORγt. IRF4 further controls Gata3 expression, whereas BATF amplifies IL-17 synthesis. The ILE HMG TFs still exert modulatory functions as NOTCH enhances expression of effector cell type–specific TFs and promotes cytokine gene expression. Conversely, TCF1 negatively regulates IFN-γ and IL-17 production, but its activity is downmodulated by TCR signaling. SOX4 and SOX5, two additional HMG TFs, modulate Th2 and Th17 cell differentiation, respectively. SOX4 inhibits GATA3 function, whereas SOX5 promotes Rorc expression. Abbreviations: CLP, clonogenic lymphoid precursor; ETP, early thymic progenitor; HMG, high-mobility group; ILC, innate lymphoid cell; ILE, innate lymphoid effector; iNKT cell, invariant NKT cell; LMPP, lymphoid-primed multipotent progenitor; RAG, recombination activating gene; RAR, retinoic acid receptor; TF, transcription factor.