Biology and therapy of inherited retinal degenerative disease: insights from mouse models
- PMID: 25650393
- PMCID: PMC4314777
- DOI: 10.1242/dmm.017913
Biology and therapy of inherited retinal degenerative disease: insights from mouse models
Abstract
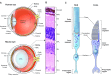
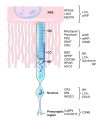
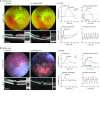
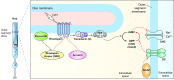
Retinal neurodegeneration associated with the dysfunction or death of photoreceptors is a major cause of incurable vision loss. Tremendous progress has been made over the last two decades in discovering genes and genetic defects that lead to retinal diseases. The primary focus has now shifted to uncovering disease mechanisms and designing treatment strategies, especially inspired by the successful application of gene therapy in some forms of congenital blindness in humans. Both spontaneous and laboratory-generated mouse mutants have been valuable for providing fundamental insights into normal retinal development and for deciphering disease pathology. Here, we provide a review of mouse models of human retinal degeneration, with a primary focus on diseases affecting photoreceptor function. We also describe models associated with retinal pigment epithelium dysfunction or synaptic abnormalities. Furthermore, we highlight the crucial role of mouse models in elucidating retinal and photoreceptor biology in health and disease, and in the assessment of novel therapeutic modalities, including gene- and stem-cell-based therapies, for retinal degenerative diseases.
Keywords: Mouse mutants; Photoreceptor; Retinal development; Retinal disease.
© 2015. Published by The Company of Biologists Ltd.
Figures
References
-
- Acland G. M., Aguirre G. D., Ray J., Zhang Q., Aleman T. S., Cideciyan A. V., Pearce-Kelling S. E., Anand V., Zeng Y., Maguire A. M., et al. (2001). Gene therapy restores vision in a canine model of childhood blindness. Nat. Genet. 28, 92–95. - PubMed
-
- Adamis A. P., Miller J. W., Bernal M. T., D’Amico D. J., Folkman J., Yeo T. K., Yeo K. T. (1994). Increased vascular endothelial growth factor levels in the vitreous of eyes with proliferative diabetic retinopathy. Am. J. Ophthalmol. 118, 445–450. - PubMed
-
- Akhmedov N. B., Piriev N. I., Chang B., Rapoport A. L., Hawes N. L., Nishina P. M., Nusinowitz S., Heckenlively J. R., Roderick T. H., Kozak C. A., et al. (2000). A deletion in a photoreceptor-specific nuclear receptor mRNA causes retinal degeneration in the rd7 mouse. Proc. Natl. Acad. Sci. USA 97, 5551–5556. - PMC - PubMed
-
- Akimoto M., Cheng H., Zhu D., Brzezinski J. A., Khanna R., Filippova E., Oh E. C., Jing Y., Linares J. L., Brooks M., et al. (2006). Targeting of GFP to newborn rods by Nrl promoter and temporal expression profiling of flow-sorted photoreceptors. Proc. Natl. Acad. Sci. USA 103, 3890–3895. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

