The selection and function of cell type-specific enhancers
- PMID: 25650801
- PMCID: PMC4517609
- DOI: 10.1038/nrm3949
The selection and function of cell type-specific enhancers
Abstract
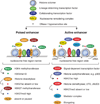
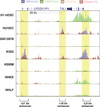
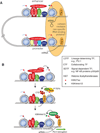
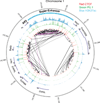
The human body contains several hundred cell types, all of which share the same genome. In metazoans, much of the regulatory code that drives cell type-specific gene expression is located in distal elements called enhancers. Although mammalian genomes contain millions of potential enhancers, only a small subset of them is active in a given cell type. Cell type-specific enhancer selection involves the binding of lineage-determining transcription factors that prime enhancers. Signal-dependent transcription factors bind to primed enhancers, which enables these broadly expressed factors to regulate gene expression in a cell type-specific manner. The expression of genes that specify cell type identity and function is associated with densely spaced clusters of active enhancers known as super-enhancers. The functions of enhancers and super-enhancers are influenced by, and affect, higher-order genomic organization.
Figures

References
-
- Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. - PubMed
-
- Lettice LA, et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12:1725–1735. - PubMed
-
- Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. - PubMed
-
- Carroll JS, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

