Identification of natural RORγ ligands that regulate the development of lymphoid cells
- PMID: 25651181
- PMCID: PMC4317570
- DOI: 10.1016/j.cmet.2015.01.004
Identification of natural RORγ ligands that regulate the development of lymphoid cells
Abstract
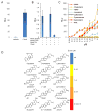
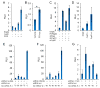
Mice deficient in the nuclear hormone receptor RORγt have defective development of thymocytes, lymphoid organs, Th17 cells, and type 3 innate lymphoid cells. RORγt binds to oxysterols derived from cholesterol catabolism, but it is not clear whether these are its natural ligands. Here, we show that sterol lipids are necessary and sufficient to drive RORγt-dependent transcription. We combined overexpression, RNAi, and genetic deletion of metabolic enzymes to study RORγ-dependent transcription. Our results are consistent with the RORγt ligand(s) being a cholesterol biosynthetic intermediate (CBI) downstream of lanosterol and upstream of zymosterol. Analysis of lipids bound to RORγ identified molecules with molecular weights consistent with CBIs. Furthermore, CBIs stabilized the RORγ ligand-binding domain and induced coactivator recruitment. Genetic deletion of metabolic enzymes upstream of the RORγt-ligand(s) affected the development of lymph nodes and Th17 cells. Our data suggest that CBIs play a role in lymphocyte development potentially through regulation of RORγt.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures



References
-
- Bai J, Uehara Y, Montell DJ. Regulation of invasive cell behavior by taiman, a Drosophila protein related to AIB1, a steroid receptor coactivator amplified in breast cancer. Cell. 2000;103:1047–1058. - PubMed
-
- Billheimer JT, Chamoun D, Esfahani M. Defective 3-ketosteroid reductase activity in a human monocyte-like cell line. J Lipid Res. 1987;28:704–709. - PubMed
-
- Bloch K. The biological synthesis of cholesterol. Science. 1965;150:19–28. - PubMed
-
- Byskov AG, Andersen CY, Nordholm L, Thogersen H, Xia G, Wassmann O, Andersen JV, Guddal E, Roed T. Chemical structure of sterols that activate oocyte meiosis. Nature. 1995;374:559–562. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

