Evaluation and improvements of clustering algorithms for detecting remote homologous protein families
- PMID: 25651949
- PMCID: PMC4339679
- DOI: 10.1186/s12859-014-0445-4
Evaluation and improvements of clustering algorithms for detecting remote homologous protein families
Abstract
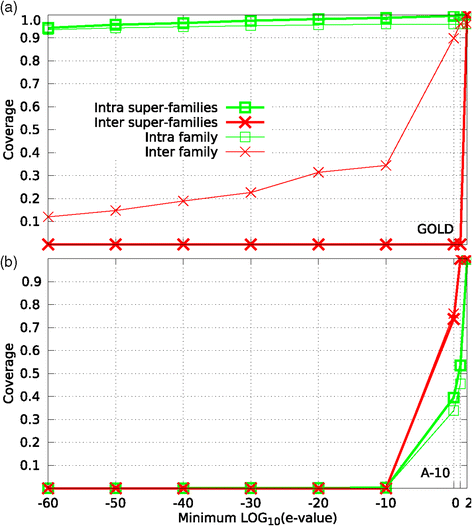
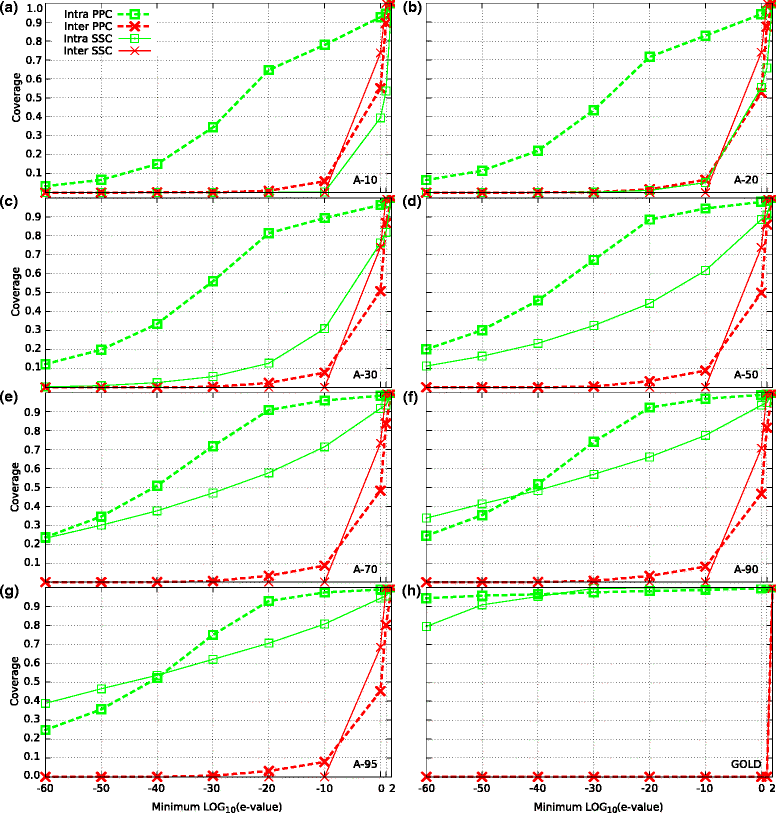
Background: An important problem in computational biology is the automatic detection of protein families (groups of homologous sequences). Clustering sequences into families is at the heart of most comparative studies dealing with protein evolution, structure, and function. Many methods have been developed for this task, and they perform reasonably well (over 0.88 of F-measure) when grouping proteins with high sequence identity. However, for highly diverged proteins the performance of these methods can be much lower, mainly because a common evolutionary origin is not deduced directly from sequence similarity. To the best of our knowledge, a systematic evaluation of clustering methods over distant homologous proteins is still lacking.
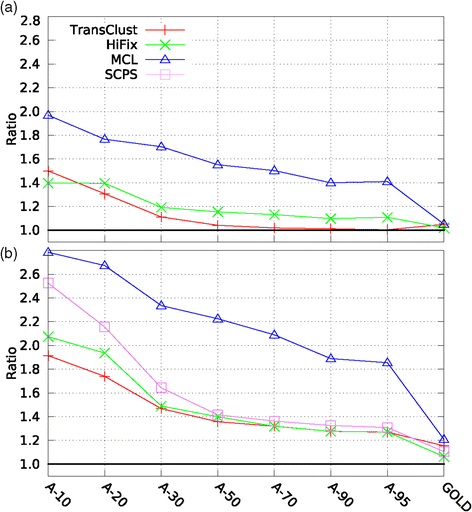
Results: We performed a comparative assessment of four clustering algorithms: Markov Clustering (MCL), Transitive Clustering (TransClust), Spectral Clustering of Protein Sequences (SCPS), and High-Fidelity clustering of protein sequences (HiFix), considering several datasets with different levels of sequence similarity. Two types of similarity measures, required by the clustering sequence methods, were used to evaluate the performance of the algorithms: the standard measure obtained from sequence-sequence comparisons, and a novel measure based on profile-profile comparisons, used here for the first time.
Conclusions: The results reveal low clustering performance for the highly divergent datasets when the standard measure was used. However, the novel measure based on profile-profile comparisons substantially improved the performance of the four methods, especially when very low sequence identity datasets were evaluated. We also performed a parameter optimization step to determine the best configuration for each clustering method. We found that TransClust clearly outperformed the other methods for most datasets. This work also provides guidelines for the practical application of clustering sequence methods aimed at detecting accurately groups of related protein sequences.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

