Fine-mapping identifies two additional breast cancer susceptibility loci at 9q31.2
- PMID: 25652398
- PMCID: PMC4406292
- DOI: 10.1093/hmg/ddv035
Fine-mapping identifies two additional breast cancer susceptibility loci at 9q31.2
Abstract
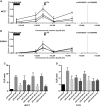
We recently identified a novel susceptibility variant, rs865686, for estrogen-receptor positive breast cancer at 9q31.2. Here, we report a fine-mapping analysis of the 9q31.2 susceptibility locus using 43 160 cases and 42 600 controls of European ancestry ascertained from 52 studies and a further 5795 cases and 6624 controls of Asian ancestry from nine studies. Single nucleotide polymorphism (SNP) rs676256 was most strongly associated with risk in Europeans (odds ratios [OR] = 0.90 [0.88-0.92]; P-value = 1.58 × 10(-25)). This SNP is one of a cluster of highly correlated variants, including rs865686, that spans ∼14.5 kb. We identified two additional independent association signals demarcated by SNPs rs10816625 (OR = 1.12 [1.08-1.17]; P-value = 7.89 × 10(-09)) and rs13294895 (OR = 1.09 [1.06-1.12]; P-value = 2.97 × 10(-11)). SNP rs10816625, but not rs13294895, was also associated with risk of breast cancer in Asian individuals (OR = 1.12 [1.06-1.18]; P-value = 2.77 × 10(-05)). Functional genomic annotation using data derived from breast cancer cell-line models indicates that these SNPs localise to putative enhancer elements that bind known drivers of hormone-dependent breast cancer, including ER-α, FOXA1 and GATA-3. In vitro analyses indicate that rs10816625 and rs13294895 have allele-specific effects on enhancer activity and suggest chromatin interactions with the KLF4 gene locus. These results demonstrate the power of dense genotyping in large studies to identify independent susceptibility variants. Analysis of associations using subjects with different ancestry, combined with bioinformatic and genomic characterisation, can provide strong evidence for the likely causative alleles and their functional basis.
© The Author 2015. Published by Oxford University Press.
Figures


References
-
- Ferlay J., Shin H.R., Bray F., Forman D., Mathers C., Parkin D.M. (2010) Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer, 127, 2893–2917. - PubMed
-
- Pharoah P.D., Antoniou A., Bobrow M., Zimmern R.L., Easton D.F., Ponder B.A. (2002) Polygenic susceptibility to breast cancer and implications for prevention. Nat. Genet., 31, 33–36. - PubMed
-
- Meijers-Heijboer H., van den Ouweland A., Klijn J., Wasielewski M., de Snoo A., Oldenburg R., Hollestelle A., Houben M., Crepin E., van Veghel-Plandsoen M., et al. (2002) Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat. Genet., 31, 55–59. - PubMed
-
- Renwick A., Thompson D., Seal S., Kelly P., Chagtai T., Ahmed M., North B., Jayatilake H., Barfoot R., Spanova K., et al. (2006) ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat. Genet., 38, 873–875. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA128978/CA/NCI NIH HHS/United States
- R01 CA176785/CA/NCI NIH HHS/United States
- P30 CA16056/CA/NCI NIH HHS/United States
- R01 CA128978/CA/NCI NIH HHS/United States
- U19 CA148065/CA/NCI NIH HHS/United States
- C1281/A12014/CRUK_/Cancer Research UK/United Kingdom
- MC_PC_14105/MRC_/Medical Research Council/United Kingdom
- CA116201/CA/NCI NIH HHS/United States
- P30 CA016056/CA/NCI NIH HHS/United States
- MR/K006215/1/MRC_/Medical Research Council/United Kingdom
- C1287/A10710/CRUK_/Cancer Research UK/United Kingdom
- R01CA64277/CA/NCI NIH HHS/United States
- UM1 CA164920/CA/NCI NIH HHS/United States
- P30 CA68485/CA/NCI NIH HHS/United States
- R37 CA070867/CA/NCI NIH HHS/United States
- 16565/CRUK_/Cancer Research UK/United Kingdom
- R01 CA092447/CA/NCI NIH HHS/United States
- CAPMC/ CIHR/Canada
- C12292/A11174/CRUK_/Cancer Research UK/United Kingdom
- R01CA148667/CA/NCI NIH HHS/United States
- 1U19 CA148065/CA/NCI NIH HHS/United States
- C5047/A10692/CRUK_/Cancer Research UK/United Kingdom
- 1U19 CA148537/CA/NCI NIH HHS/United States
- R01 CA100374/CA/NCI NIH HHS/United States
- C490/A10124/CRUK_/Cancer Research UK/United Kingdom
- U01 CA116167/CA/NCI NIH HHS/United States
- R01CA100374/CA/NCI NIH HHS/United States
- R01 CA064277/CA/NCI NIH HHS/United States
- CA116167/CA/NCI NIH HHS/United States
- CA176785/CA/NCI NIH HHS/United States
- U19 CA148537/CA/NCI NIH HHS/United States
- R01 CA116167/CA/NCI NIH HHS/United States
- R01 CA148667/CA/NCI NIH HHS/United States
- R37CA70867/CA/NCI NIH HHS/United States
- P50 CA116201/CA/NCI NIH HHS/United States
- C1287/A12014/CRUK_/Cancer Research UK/United Kingdom
- R01 CA063464/CA/NCI NIH HHS/United States
- P30 CA015083/CA/NCI NIH HHS/United States
- C5047/A15007/CRUK_/Cancer Research UK/United Kingdom
- 16563/CRUK_/Cancer Research UK/United Kingdom
- U01 CA063464/CA/NCI NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- R01 CA077398/CA/NCI NIH HHS/United States
- R01 CA054281/CA/NCI NIH HHS/United States
- R01 CA132839/CA/NCI NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- R01 CA77398/CA/NCI NIH HHS/United States
- 1U19 CA148112/CA/NCI NIH HHS/United States
- U19 CA148112/CA/NCI NIH HHS/United States
- UM1CA164920/CA/NCI NIH HHS/United States
- C5047A8384/CRUK_/Cancer Research UK/United Kingdom
- U01 CA098758/CA/NCI NIH HHS/United States
- N01 CN25403/CN/NCI NIH HHS/United States
- N01 CN025403/CA/NCI NIH HHS/United States
- C8197/A16565/CRUK_/Cancer Research UK/United Kingdom
- UM1 CA182910/CA/NCI NIH HHS/United States
- C1287/A10118/CRUK_/Cancer Research UK/United Kingdom
- 001/WHO_/World Health Organization/International
- R37 CA054281/CA/NCI NIH HHS/United States
- 16561/CRUK_/Cancer Research UK/United Kingdom
- 10124/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

