Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel
- PMID: 25653097
- PMCID: PMC4338501
- DOI: 10.1038/ncomms4934
Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel
Abstract
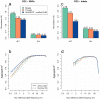
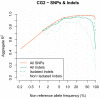
A major use of the 1000 Genomes Project (1000 GP) data is genotype imputation in genome-wide association studies (GWAS). Here we develop a method to estimate haplotypes from low-coverage sequencing data that can take advantage of single-nucleotide polymorphism (SNP) microarray genotypes on the same samples. First the SNP array data are phased to build a backbone (or 'scaffold') of haplotypes across each chromosome. We then phase the sequence data 'onto' this haplotype scaffold. This approach can take advantage of relatedness between sequenced and non-sequenced samples to improve accuracy. We use this method to create a new 1000 GP haplotype reference set for use by the human genetic community. Using a set of validation genotypes at SNP and bi-allelic indels we show that these haplotypes have lower genotype discordance and improved imputation performance into downstream GWAS samples, especially at low-frequency variants.
Figures

References
-
- Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 2010;11:499–511. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

